[English] 日本語
 Yorodumi
Yorodumi- EMDB-27654: A subtomogram average of H. neapolitanus Rubisco within alpha-car... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | A subtomogram average of H. neapolitanus Rubisco within alpha-carboxysomes | ||||||||||||
 Map data Map data | Subtomogram average of Rubisco within H. neapolitanus alpha-carboxysome. Contour level for best display of interior. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Carbon fixation / alpha-carboxysome / enzyme / LYASE | ||||||||||||
| Biological species |  Halothiobacillus neapolitanus (bacteria) Halothiobacillus neapolitanus (bacteria) | ||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 4.5 Å | ||||||||||||
 Authors Authors | Metskas LA / Blikstad C / Laughlin T / Savage DF / Jensen GJ | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Rubisco forms a lattice inside alpha-carboxysomes. Authors: Lauren Ann Metskas / Davi Ortega / Luke M Oltrogge / Cecilia Blikstad / Derik R Lovejoy / Thomas G Laughlin / David F Savage / Grant J Jensen /   Abstract: Despite the importance of microcompartments in prokaryotic biology and bioengineering, structural heterogeneity has prevented a complete understanding of their architecture, ultrastructure, and ...Despite the importance of microcompartments in prokaryotic biology and bioengineering, structural heterogeneity has prevented a complete understanding of their architecture, ultrastructure, and spatial organization. Here, we employ cryo-electron tomography to image α-carboxysomes, a pseudo-icosahedral microcompartment responsible for carbon fixation. We have solved a high-resolution subtomogram average of the Rubisco cargo inside the carboxysome, and determined the arrangement of the enzyme. We find that the H. neapolitanus Rubisco polymerizes in vivo, mediated by the small Rubisco subunit. These fibrils can further pack to form a lattice with six-fold pseudo-symmetry. This arrangement preserves freedom of motion and accessibility around the Rubisco active site and the binding sites for two other carboxysome proteins, CsoSCA (a carbonic anhydrase) and the disordered CsoS2, even at Rubisco concentrations exceeding 800 μM. This characterization of Rubisco cargo inside the α-carboxysome provides insight into the balance between order and disorder in microcompartment organization. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27654.map.gz emd_27654.map.gz | 5.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27654-v30.xml emd-27654-v30.xml emd-27654.xml emd-27654.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27654_fsc.xml emd_27654_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_27654.png emd_27654.png | 81.9 KB | ||
| Masks |  emd_27654_msk_1.map emd_27654_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27654.cif.gz emd-27654.cif.gz | 4.3 KB | ||
| Others |  emd_27654_half_map_1.map.gz emd_27654_half_map_1.map.gz emd_27654_half_map_2.map.gz emd_27654_half_map_2.map.gz | 12.7 MB 14.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27654 http://ftp.pdbj.org/pub/emdb/structures/EMD-27654 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27654 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27654 | HTTPS FTP |
-Validation report
| Summary document |  emd_27654_validation.pdf.gz emd_27654_validation.pdf.gz | 831 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27654_full_validation.pdf.gz emd_27654_full_validation.pdf.gz | 830.5 KB | Display | |
| Data in XML |  emd_27654_validation.xml.gz emd_27654_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_27654_validation.cif.gz emd_27654_validation.cif.gz | 16 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27654 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27654 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27654 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27654 | HTTPS FTP |
-Related structure data
| EM raw data |  EMPIAR-11125 (Title: Cryo electron tomography of H. neapolitanus alpha-carboxysomes EMPIAR-11125 (Title: Cryo electron tomography of H. neapolitanus alpha-carboxysomesData size: 483.4 Data #1: Reconstructed sample tomograms of purified alpha-carboxysomes, plus frames [reconstructed volumes] Data #2: Reconstructed sample tomograms of wild-type H. neapolitanus cells, plus frames [reconstructed volumes] Data #3: Complete set of subtomograms for Rubisco subtomogram average [picked particles - multiframe - unprocessed]) |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27654.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27654.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average of Rubisco within H. neapolitanus alpha-carboxysome. Contour level for best display of interior. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.104 Å | ||||||||||||||||||||||||||||||||||||
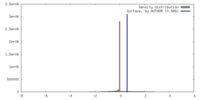
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27654_msk_1.map emd_27654_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
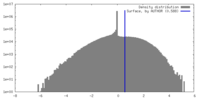
| Density Histograms |
-Half map: Half-map 1, unfiltered
| File | emd_27654_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1, unfiltered | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2, unfiltered
| File | emd_27654_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2, unfiltered | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rubisco within the alpha-carboxysome
| Entire | Name: Rubisco within the alpha-carboxysome |
|---|---|
| Components |
|
-Supramolecule #1: Rubisco within the alpha-carboxysome
| Supramolecule | Name: Rubisco within the alpha-carboxysome / type: organelle_or_cellular_component / ID: 1 / Parent: 0 Details: Subtomogram average of Rubisco within carboxysomes purified from H. neapolitanus |
|---|---|
| Source (natural) | Organism:  Halothiobacillus neapolitanus (bacteria) / Location in cell: Alpha-carboxysome Halothiobacillus neapolitanus (bacteria) / Location in cell: Alpha-carboxysome |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | Purified alpha-carboxysome |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average electron dose: 2.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)