[English] 日本語
 Yorodumi
Yorodumi- EMDB-27633: Cryo-EM structure of the 5HT2C receptor (INI isoform) bound to lo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the 5HT2C receptor (INI isoform) bound to lorcaserin | |||||||||
 Map data Map data | deepEMhancer sharp map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of corticotropin-releasing hormone secretion / 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding / Gq/11-coupled serotonin receptor activity / phospholipase C-activating serotonin receptor signaling pathway / positive regulation of phosphatidylinositol biosynthetic process / G protein-coupled serotonin receptor signaling pathway / regulation of appetite / G protein-coupled serotonin receptor complex / regulation of nervous system process / Serotonin receptors ...regulation of corticotropin-releasing hormone secretion / 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding / Gq/11-coupled serotonin receptor activity / phospholipase C-activating serotonin receptor signaling pathway / positive regulation of phosphatidylinositol biosynthetic process / G protein-coupled serotonin receptor signaling pathway / regulation of appetite / G protein-coupled serotonin receptor complex / regulation of nervous system process / Serotonin receptors / serotonin receptor signaling pathway / serotonin binding / G protein-coupled serotonin receptor activity / feeding behavior / cGMP-mediated signaling / neurotransmitter receptor activity / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / behavioral fear response / positive regulation of fat cell differentiation / release of sequestered calcium ion into cytosol / positive regulation of calcium-mediated signaling / locomotory behavior / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / intracellular calcium ion homeostasis / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / GTPase binding / retina development in camera-type eye / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / chemical synaptic transmission / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / positive regulation of ERK1 and ERK2 cascade / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / dendrite / synapse / protein-containing complex binding / signal transduction / extracellular exosome / identical protein binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
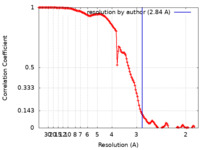
| Method | single particle reconstruction / cryo EM / Resolution: 2.84 Å | |||||||||
 Authors Authors | Gumpper RH / Fay JF / Roth BL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: Molecular insights into the regulation of constitutive activity by RNA editing of 5HT serotonin receptors. Authors: Ryan H Gumpper / Jonathan F Fay / Bryan L Roth /  Abstract: RNA editing is a process by which post-transcriptional changes of mRNA nucleotides alter protein function through modification of the amino acid content. The 5HT serotonin receptor, which undergoes ...RNA editing is a process by which post-transcriptional changes of mRNA nucleotides alter protein function through modification of the amino acid content. The 5HT serotonin receptor, which undergoes 32 distinct RNA-editing events leading to 24 protein isoforms, is a notable example of this process. These 5HT isoforms display differences in constitutive activity, agonist/inverse agonist potencies, and efficacies. To elucidate the molecular mechanisms responsible for these effects of RNA editing, we present four active-state 5HT-transducer-coupled structures of three representative isoforms (INI, VGV, and VSV) with the selective drug lorcaserin (Belviq) and the classic psychedelic psilocin. We also provide a comprehensive analysis of agonist activation and constitutive activity across all 24 protein isoforms. Collectively, these findings reveal a unique hydrogen-bonding network located on intracellular loop 2 that is subject to RNA editing, which differentially affects GPCR constitutive and agonist signaling activities. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27633.map.gz emd_27633.map.gz | 78.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27633-v30.xml emd-27633-v30.xml emd-27633.xml emd-27633.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27633_fsc.xml emd_27633_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_27633.png emd_27633.png | 72.3 KB | ||
| Filedesc metadata |  emd-27633.cif.gz emd-27633.cif.gz | 6.4 KB | ||
| Others |  emd_27633_additional_1.map.gz emd_27633_additional_1.map.gz emd_27633_half_map_1.map.gz emd_27633_half_map_1.map.gz emd_27633_half_map_2.map.gz emd_27633_half_map_2.map.gz | 85.2 MB 84.6 MB 84.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27633 http://ftp.pdbj.org/pub/emdb/structures/EMD-27633 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27633 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27633 | HTTPS FTP |
-Validation report
| Summary document |  emd_27633_validation.pdf.gz emd_27633_validation.pdf.gz | 726.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27633_full_validation.pdf.gz emd_27633_full_validation.pdf.gz | 725.9 KB | Display | |
| Data in XML |  emd_27633_validation.xml.gz emd_27633_validation.xml.gz | 18.9 KB | Display | |
| Data in CIF |  emd_27633_validation.cif.gz emd_27633_validation.cif.gz | 25 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27633 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27633 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27633 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27633 | HTTPS FTP |
-Related structure data
| Related structure data |  8dpfMC  8dpgC  8dphC  8dpiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27633.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27633.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMhancer sharp map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||
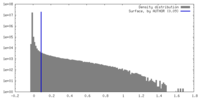
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Bfactor sharp map
| File | emd_27633_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bfactor sharp map | ||||||||||||
| Projections & Slices |
| ||||||||||||
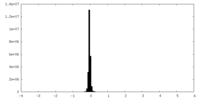
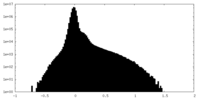
| Density Histograms |
-Half map: halfmap A
| File | emd_27633_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap A | ||||||||||||
| Projections & Slices |
| ||||||||||||
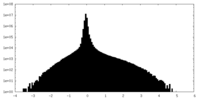
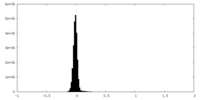
| Density Histograms |
-Half map: halfmap B
| File | emd_27633_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap B | ||||||||||||
| Projections & Slices |
| ||||||||||||
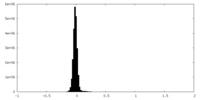
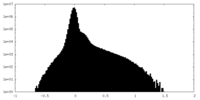
| Density Histograms |
- Sample components
Sample components
-Entire : 5-ht2c heterotrimeric complex (INI isoform)
| Entire | Name: 5-ht2c heterotrimeric complex (INI isoform) |
|---|---|
| Components |
|
-Supramolecule #1: 5-ht2c heterotrimeric complex (INI isoform)
| Supramolecule | Name: 5-ht2c heterotrimeric complex (INI isoform) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: 5-hydroxytryptamine receptor 2C
| Macromolecule | Name: 5-hydroxytryptamine receptor 2C / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.876066 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVNLRNAVHS FLVHLIGLLV WQCDISVSPV AAIVTDIFNT SDGGRFKFPD GVQNWPALSI VIIIIMTIGG NILVIMAVSM EKKLHNATN YFLMSLAIAD MLVGLLVMPL SLLAILYDYV WPLPRYLCPV WISLDVLFST ASIMHLCAIS LDRYVAIRNP I EHSRFNSR ...String: MVNLRNAVHS FLVHLIGLLV WQCDISVSPV AAIVTDIFNT SDGGRFKFPD GVQNWPALSI VIIIIMTIGG NILVIMAVSM EKKLHNATN YFLMSLAIAD MLVGLLVMPL SLLAILYDYV WPLPRYLCPV WISLDVLFST ASIMHLCAIS LDRYVAIRNP I EHSRFNSR TKAIMKIAIV WAISIGVSVP IPVIGLRDEE KVFVNNTTCV LNDPNFVLIG SFVAFFIPLT IMVITYCLTI YV LRRQALM LLHGHTEEPP GLSLDFLKCC KRNTAEEENS ANPNQDQNAR RRKKKERRPR GTMQAINNER KASKVLGIVF FVF LIMWCP FFITNILSVL CEKSCNQKLM EKLLNVFVWI GYVCSGINPL VYTLFNKIYR RAFSNYLRCN YKVEKKPPVR QIPR VAATA LSGRELNVNI YRHTNEPVIE KASDNEPGIE MQVENLELPV NPSSVVSERI SSV UniProtKB: 5-hydroxytryptamine receptor 2C |
-Macromolecule #2: G-alpha subunit q (Gi2-mini-Gq chimeric)
| Macromolecule | Name: G-alpha subunit q (Gi2-mini-Gq chimeric) / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.084832 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSTVSAEDK AAAERSKMID KNLREDGEKA RRTLRLLLLG ADNSGKSTIV KQMRILHGGS GGSGGTSGIF ETKFQVDKVN FHMFDVGGQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA ...String: MGSTVSAEDK AAAERSKMID KNLREDGEKA RRTLRLLLLG ADNSGKSTIV KQMRILHGGS GGSGGTSGIF ETKFQVDKVN FHMFDVGGQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA RYTTPEDATP EPGEDPRVTR AKYFIRKEFV DISTASGDGR HICYPHFTCA VDTENARRIF NDCKDIILQM NL REYNLV |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Antibody fragment scFv16
| Macromolecule | Name: Antibody fragment scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.668922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAALEVLFQ GPHHHHHHHH |
-Macromolecule #6: (1R)-8-chloro-1-methyl-2,3,4,5-tetrahydro-1H-3-benzazepine
| Macromolecule | Name: (1R)-8-chloro-1-methyl-2,3,4,5-tetrahydro-1H-3-benzazepine type: ligand / ID: 6 / Number of copies: 1 / Formula: T4U |
|---|---|
| Molecular weight | Theoretical: 195.689 Da |
| Chemical component information | 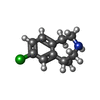 ChemComp-T4U: |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 49.6 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.551 µm / Nominal defocus min: 0.421 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)