+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of S. epidermidis Cas10-Csm bound to target RNA | |||||||||
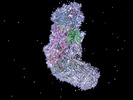
 Map data Map data | Cas10-Csm complex bound to crRNA and target RNA. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / Cas10 / Type III / Csm / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationdefense response to virus / endonuclease activity / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Paraan M / Stagg SM / Dunkle JA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS One / Year: 2023 Journal: PLoS One / Year: 2023Title: The structure of a Type III-A CRISPR-Cas effector complex reveals conserved and idiosyncratic contacts to target RNA and crRNA among Type III-A systems. Authors: Mohammadreza Paraan / Mohamed Nasef / Lucy Chou-Zheng / Sarah A Khweis / Allyn J Schoeffler / Asma Hatoum-Aslan / Scott M Stagg / Jack A Dunkle /  Abstract: Type III CRISPR-Cas systems employ multiprotein effector complexes bound to small CRISPR RNAs (crRNAs) to detect foreign RNA transcripts and elicit a complex immune response that leads to the ...Type III CRISPR-Cas systems employ multiprotein effector complexes bound to small CRISPR RNAs (crRNAs) to detect foreign RNA transcripts and elicit a complex immune response that leads to the destruction of invading RNA and DNA. Type III systems are among the most widespread in nature, and emerging interest in harnessing these systems for biotechnology applications highlights the need for detailed structural analyses of representatives from diverse organisms. We performed cryo-EM reconstructions of the Type III-A Cas10-Csm effector complex from S. epidermidis bound to an intact, cognate target RNA and identified two oligomeric states, a 276 kDa complex and a 318 kDa complex. 3.1 Å density for the well-ordered 276 kDa complex allowed construction of atomic models for the Csm2, Csm3, Csm4 and Csm5 subunits within the complex along with the crRNA and target RNA. We also collected small-angle X-ray scattering data which was consistent with the 276 kDa Cas10-Csm architecture we identified. Detailed comparisons between the S. epidermidis Cas10-Csm structure and the well-resolved bacterial (S. thermophilus) and archaeal (T. onnurineus) Cas10-Csm structures reveal differences in how the complexes interact with target RNA and crRNA which are likely to have functional ramifications. These structural comparisons shed light on the unique features of Type III-A systems from diverse organisms and will assist in improving biotechnologies derived from Type III-A effector complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27593.map.gz emd_27593.map.gz | 230.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27593-v30.xml emd-27593-v30.xml emd-27593.xml emd-27593.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27593.png emd_27593.png | 71.8 KB | ||
| Filedesc metadata |  emd-27593.cif.gz emd-27593.cif.gz | 6.5 KB | ||
| Others |  emd_27593_half_map_1.map.gz emd_27593_half_map_1.map.gz emd_27593_half_map_2.map.gz emd_27593_half_map_2.map.gz | 226.8 MB 226.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27593 http://ftp.pdbj.org/pub/emdb/structures/EMD-27593 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27593 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27593 | HTTPS FTP |
-Validation report
| Summary document |  emd_27593_validation.pdf.gz emd_27593_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27593_full_validation.pdf.gz emd_27593_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_27593_validation.xml.gz emd_27593_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_27593_validation.cif.gz emd_27593_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27593 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27593 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27593 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27593 | HTTPS FTP |
-Related structure data
| Related structure data |  8do6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27593.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27593.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cas10-Csm complex bound to crRNA and target RNA. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.846 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_27593_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_27593_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cas10-Csm complex bound to target RNA
| Entire | Name: Cas10-Csm complex bound to target RNA |
|---|---|
| Components |
|
-Supramolecule #1: Cas10-Csm complex bound to target RNA
| Supramolecule | Name: Cas10-Csm complex bound to target RNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: CRISPR system Cms endoribonuclease Csm3
| Macromolecule | Name: CRISPR system Cms endoribonuclease Csm3 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A |
| Molecular weight | Theoretical: 24.033975 KDa |
| Recombinant expression | Organism:  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) |
| Sequence | String: MYSKIKISGT IEVVTGLHIG GGGESSMIGA IDSPVVRDLQ TKLPIIPGSS IKGKMRNLLA KHFGLKMKQE SHNQDDERVL RLFGSSEKG NIQRARLQIS DAFFSEKTKE HFAQNDIAYT ETKFENTINR LTAVANPRQI ERVTRGSEFD FVFIYNVDEE S QVEDDFEN ...String: MYSKIKISGT IEVVTGLHIG GGGESSMIGA IDSPVVRDLQ TKLPIIPGSS IKGKMRNLLA KHFGLKMKQE SHNQDDERVL RLFGSSEKG NIQRARLQIS DAFFSEKTKE HFAQNDIAYT ETKFENTINR LTAVANPRQI ERVTRGSEFD FVFIYNVDEE S QVEDDFEN IEKAIHLLEN DYLGGGGTRG NGRIQFKDTN IETVVGEYDS TNLKIK UniProtKB: CRISPR system Cms endoribonuclease Csm3 |
-Macromolecule #4: CRISPR system Cms protein Csm2
| Macromolecule | Name: CRISPR system Cms protein Csm2 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) |
| Molecular weight | Theoretical: 16.809471 KDa |
| Recombinant expression | Organism:  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) |
| Sequence | String: MILAKTKSGK TIDLTFAHEV VKSNVKNVKD RKGKEKQVLF NGLTTSKLRN LMEQVNRLYT IAFNSNEDQL NEEFIDELEY LKIKFYYEA GREKSVDEFL KKTLMFPIID RVIKKESKKF FLDYCKYFEA LVAYAKYYQK ED UniProtKB: UNIPROTKB: A0A8G7QML1 |
-Macromolecule #5: CRISPR system Cms protein Csm4
| Macromolecule | Name: CRISPR system Cms protein Csm4 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A |
| Molecular weight | Theoretical: 34.551938 KDa |
| Recombinant expression | Organism:  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) |
| Sequence | String: MTLATKVFKL SFKTPVHFGK KRLSDGEMTI TADTLFSALF IETLQLGKDT DWLLNDLIIS DTFPYENELY YLPKPLIKID SKEEDNHKA FKKLKYVPVH HYNQYLNGEL SAEDATDLND IFNIGYFSLQ TKVSLIAQET DSSADSEPYS VGTFTFEPEA G LYFIAKGS ...String: MTLATKVFKL SFKTPVHFGK KRLSDGEMTI TADTLFSALF IETLQLGKDT DWLLNDLIIS DTFPYENELY YLPKPLIKID SKEEDNHKA FKKLKYVPVH HYNQYLNGEL SAEDATDLND IFNIGYFSLQ TKVSLIAQET DSSADSEPYS VGTFTFEPEA G LYFIAKGS EETLDHLNNI MTALQYSGLG GKRNAGYGQF EYEIINNQQL SKLLNQNGKH SILLSTAMAK KEEIESALKE AR YILTKRS GFVQSTNYSE MLVKKSDFYS FSSGSVFKNI FNGDIFNVGH NGKHPVYRYA KPLWLEV UniProtKB: CRISPR system Cms protein Csm4 |
-Macromolecule #6: CRISPR system Cms protein Csm5
| Macromolecule | Name: CRISPR system Cms protein Csm5 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A |
| Molecular weight | Theoretical: 39.449125 KDa |
| Recombinant expression | Organism:  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) |
| Sequence | String: MTIKNYEVVI KTLGPIHIGS GQVMKKQDYI YDFYNSKVYM INGNKLVKFL KRKNLLYTYQ NFLRYPPKNP RENGLKDYLD AQNVKQSEW EAFVSYSEKV NQGKKYGNTR PKPLNDLHLM VRDGQNKVYL PGSSIKGAIK TTLVSKYNNE KNKDIYSKIK V SDSKPIDE ...String: MTIKNYEVVI KTLGPIHIGS GQVMKKQDYI YDFYNSKVYM INGNKLVKFL KRKNLLYTYQ NFLRYPPKNP RENGLKDYLD AQNVKQSEW EAFVSYSEKV NQGKKYGNTR PKPLNDLHLM VRDGQNKVYL PGSSIKGAIK TTLVSKYNNE KNKDIYSKIK V SDSKPIDE SNLAIYQKID INKSEKSMPL YRECIDVNTE IKFKLTIEDE IYSINEIEQS IQDFYKNYYD KWLVGFKETK GG RRFALEG GIPDVLNQNI LFLGAGTGFV SKTTHYQLKN RKQAKQDSFE ILTKKFRGTY GKMKEIPSNV PVALKGTTNQ SRH TSYQQG MCKVSFQELN NEVL UniProtKB: CRISPR system Cms protein Csm5 |
-Macromolecule #2: crRNA
| Macromolecule | Name: crRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) |
| Molecular weight | Theoretical: 11.895168 KDa |
| Sequence | String: ACGAGAACAC GUAUGCCGAA GUAUAUAAAU CAUCAGU |
-Macromolecule #3: Target RNA
| Macromolecule | Name: Target RNA / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) |
| Molecular weight | Theoretical: 13.599979 KDa |
| Sequence | String: CUUUGUACUG AUGAUUUAUA UACUUCGGCA UACGUUCUCU AAA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Component:
| ||||||
|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 78.0 K / Max: 92.0 K |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 5000 / Average exposure time: 1.0 sec. / Average electron dose: 44.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 1.5 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 81000 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 0.001 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)