[English] 日本語
 Yorodumi
Yorodumi- EMDB-27513: Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with Fab 4A8 | |||||||||
 Map data Map data | structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with Fab 4A8 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
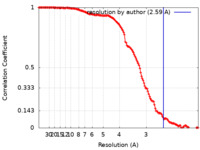
| Method | single particle reconstruction / cryo EM / Resolution: 2.59 Å | |||||||||
 Authors Authors | Zhu X / Mannar D / Saville JW / Srivastava SS / Berezuk AM / Zhou S / Tuttle KS / Subramaniam S | |||||||||
| Funding support |  Canada, 2 items Canada, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization. Authors: Dhiraj Mannar / James W Saville / Zehua Sun / Xing Zhu / Michelle M Marti / Shanti S Srivastava / Alison M Berezuk / Steven Zhou / Katharine S Tuttle / Michele D Sobolewski / Andrew Kim / ...Authors: Dhiraj Mannar / James W Saville / Zehua Sun / Xing Zhu / Michelle M Marti / Shanti S Srivastava / Alison M Berezuk / Steven Zhou / Katharine S Tuttle / Michele D Sobolewski / Andrew Kim / Benjamin R Treat / Priscila Mayrelle Da Silva Castanha / Jana L Jacobs / Simon M Barratt-Boyes / John W Mellors / Dimiter S Dimitrov / Wei Li / Sriram Subramaniam /   Abstract: Mutations in the spike glycoproteins of SARS-CoV-2 variants of concern have independently been shown to enhance aspects of spike protein fitness. Here, we describe an antibody fragment (V ab6) that ...Mutations in the spike glycoproteins of SARS-CoV-2 variants of concern have independently been shown to enhance aspects of spike protein fitness. Here, we describe an antibody fragment (V ab6) that neutralizes all major variants including the recently emerged BA.1 and BA.2 Omicron subvariants, with a unique mode of binding revealed by cryo-EM studies. Further, we provide a comparative analysis of the mutational effects within previously emerged variant spikes and identify the structural role of mutations within the NTD and RBD in evading antibody neutralization. Our analysis shows that the highly mutated Gamma N-terminal domain exhibits considerable structural rearrangements, partially explaining its decreased neutralization by convalescent sera. Our results provide mechanistic insights into the structural, functional, and antigenic consequences of SARS-CoV-2 spike mutations and highlight a spike protein vulnerability that may be exploited to achieve broad protection against circulating variants. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27513.map.gz emd_27513.map.gz | 123 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27513-v30.xml emd-27513-v30.xml emd-27513.xml emd-27513.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27513_fsc.xml emd_27513_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_27513.png emd_27513.png | 66 KB | ||
| Others |  emd_27513_half_map_1.map.gz emd_27513_half_map_1.map.gz emd_27513_half_map_2.map.gz emd_27513_half_map_2.map.gz | 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27513 http://ftp.pdbj.org/pub/emdb/structures/EMD-27513 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27513 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27513 | HTTPS FTP |
-Validation report
| Summary document |  emd_27513_validation.pdf.gz emd_27513_validation.pdf.gz | 922.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27513_full_validation.pdf.gz emd_27513_full_validation.pdf.gz | 922.1 KB | Display | |
| Data in XML |  emd_27513_validation.xml.gz emd_27513_validation.xml.gz | 19.4 KB | Display | |
| Data in CIF |  emd_27513_validation.cif.gz emd_27513_validation.cif.gz | 25.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27513 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27513 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27513 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27513 | HTTPS FTP |
-Related structure data
| Related structure data |  8dliC  8dljC  8dlkC  8dllC  8dlmC  8dlnC  8dloC  8dlpC  8dlqC  8dlrC  8dlsC  8dltC  8dluC  8dlvC  8dlwC  8dlxC  8dlyC  8dlzC  8dm0C C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27513.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27513.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with Fab 4A8 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_27513_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_27513_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Gamma (P.1) spike protein in complex with Fab 4A8
| Entire | Name: SARS-CoV-2 Gamma (P.1) spike protein in complex with Fab 4A8 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Gamma (P.1) spike protein in complex with Fab 4A8
| Supramolecule | Name: SARS-CoV-2 Gamma (P.1) spike protein in complex with Fab 4A8 type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: SARS-CoV-2 Gamma (P.1) spike protein
| Supramolecule | Name: SARS-CoV-2 Gamma (P.1) spike protein / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Fab 4A8
| Supramolecule | Name: Fab 4A8 / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: SARS-CoV-2 Gamma (P.1) spike protein
| Macromolecule | Name: SARS-CoV-2 Gamma (P.1) spike protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MFVFLVLLPL VSSQCVNFTN RTQLPSAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNYPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY ...String: MFVFLVLLPL VSSQCVNFTN RTQLPSAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNYPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY VSQPFLMDLE GKQGNFKNLS EFVFKNIDGY FKIYSKHTPI NLVRDLPQGF SALEPLVDLP IGINITRFQT LLALHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQI APGQTGTIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVKGFNCYF PLQSYGFQPT YGVGYQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLTGTG VLTESNKKFL PFQQFGRDIA DTTDAVRDPQ TLEILDITPC SFGGVSVITP GTNTSNQVAV LYQGVNCTEV PVAIHADQLT PTWRVYSTGS NVFQTRAGCL IGAEYVNNSY ECDIPIGAGI CASYQTQTNS PGSASSVASQ SIIAYTMSLG AENSVAYSNN SIAIPTNFTI SVTTEILPVS MTKTSVDCTM YICGDSTECS NLLLQYGSFC TQLNRALTGI AVEQDKNTQE VFAQVKQIYK TPPIKDFGGF NFSQILPDPS KPSKRSPIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFGA GPALQIPFPM QMAYRFNGIG VTQNVLYENQ KLIANQFNSA IGKIQDSLSS TPSALGKLQD VVNQNAQALN TLVKQLSSNF GAISSVLNDI LSRLDPPEAE VQIDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAAIKMS ECVLGQSKRV DFCGKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIVNNTVYDP LQPELDSFKE ELDKYFKNHT SPDVDLGDIS GINASFVNIQ KEIDRLNEVA KNLNESLIDL QELGKYEQGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGRS LEVLFQGPGH HHHHHHHSAW SHPQFEKGGG SGGGGSGGSA WSHPQFEK |
-Macromolecule #2: Fab 4A8 heavy chain
| Macromolecule | Name: Fab 4A8 heavy chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDWTWRVFCL LAVAPGAHSE VQLVESGAEV KKPGASVKVS CKVSGYTLTE LSMHWVRQAP GKGLEWMGGF DPEDGETMYA QKFQGRVTMT EDTSTDTAYM ELSSLRSEDT AVYYCATSTA VAGTPDLFDY YYGMDVWGQG TTVTVSSAST KGPSVFPLAP SSKSTSGGTA ...String: MDWTWRVFCL LAVAPGAHSE VQLVESGAEV KKPGASVKVS CKVSGYTLTE LSMHWVRQAP GKGLEWMGGF DPEDGETMYA QKFQGRVTMT EDTSTDTAYM ELSSLRSEDT AVYYCATSTA VAGTPDLFDY YYGMDVWGQG TTVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKKVEPKSC GSHHHHHH |
-Macromolecule #3: Fab 4A8 light chain
| Macromolecule | Name: Fab 4A8 light chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVLQTQVFIS LLLWISGAYG EIVMTQSPLS SPVTLGQPAS ISCRSSQSLV HSDGNTYLSW LQQRPGQPPR LLIYKISNRF SGVPDRFSGS GAGTDFTLKI SRVEAEDVGV YYCTQATQFP YTFGQGTKVD IKGQPKANPT VTLFPPSSEE LQANKATLVC LISDFYPGAV ...String: MVLQTQVFIS LLLWISGAYG EIVMTQSPLS SPVTLGQPAS ISCRSSQSLV HSDGNTYLSW LQQRPGQPPR LLIYKISNRF SGVPDRFSGS GAGTDFTLKI SRVEAEDVGV YYCTQATQFP YTFGQGTKVD IKGQPKANPT VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADGSP VKAGVETTKP SKQSNNKYAA SSYLSLTPEQ WKSHRSYSCQ VTHEGSTVEK TVAPTECS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)