[English] 日本語
 Yorodumi
Yorodumi- EMDB-27503: Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2 | |||||||||
 Map data Map data | structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / glycoprotein / fusion protein / viral protein / Alpha / B.1.1.7 / ACE2 / Viral Protein-Hydrolase complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / positive regulation of gap junction assembly / regulation of cardiac conduction / regulation of vasoconstriction ...positive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / positive regulation of gap junction assembly / regulation of cardiac conduction / regulation of vasoconstriction / peptidyl-dipeptidase activity / maternal process involved in female pregnancy / angiotensin maturation / Metabolism of Angiotensinogen to Angiotensins / negative regulation of signaling receptor activity / carboxypeptidase activity / Attachment and Entry / positive regulation of cardiac muscle contraction / viral life cycle / regulation of cytokine production / blood vessel diameter maintenance / negative regulation of smooth muscle cell proliferation / brush border membrane / regulation of transmembrane transporter activity / cilium / negative regulation of ERK1 and ERK2 cascade / metallopeptidase activity / positive regulation of reactive oxygen species metabolic process / endocytic vesicle membrane / virus receptor activity / regulation of cell population proliferation / regulation of inflammatory response / endopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Potential therapeutics for SARS / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / membrane fusion / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / receptor ligand activity / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / symbiont entry into host cell / membrane raft / apical plasma membrane / endoplasmic reticulum lumen / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / cell surface / extracellular space / extracellular exosome / zinc ion binding / extracellular region / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
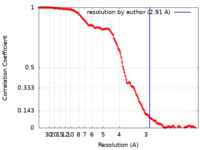
| Method | single particle reconstruction / cryo EM / Resolution: 2.91 Å | |||||||||
 Authors Authors | Zhu X / Mannar D / Saville JW / Srivastava SS / Berezuk AM / Zhou S / Tuttle KS / Subramaniam S | |||||||||
| Funding support |  Canada, 2 items Canada, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization. Authors: Dhiraj Mannar / James W Saville / Zehua Sun / Xing Zhu / Michelle M Marti / Shanti S Srivastava / Alison M Berezuk / Steven Zhou / Katharine S Tuttle / Michele D Sobolewski / Andrew Kim / ...Authors: Dhiraj Mannar / James W Saville / Zehua Sun / Xing Zhu / Michelle M Marti / Shanti S Srivastava / Alison M Berezuk / Steven Zhou / Katharine S Tuttle / Michele D Sobolewski / Andrew Kim / Benjamin R Treat / Priscila Mayrelle Da Silva Castanha / Jana L Jacobs / Simon M Barratt-Boyes / John W Mellors / Dimiter S Dimitrov / Wei Li / Sriram Subramaniam /   Abstract: Mutations in the spike glycoproteins of SARS-CoV-2 variants of concern have independently been shown to enhance aspects of spike protein fitness. Here, we describe an antibody fragment (V ab6) that ...Mutations in the spike glycoproteins of SARS-CoV-2 variants of concern have independently been shown to enhance aspects of spike protein fitness. Here, we describe an antibody fragment (V ab6) that neutralizes all major variants including the recently emerged BA.1 and BA.2 Omicron subvariants, with a unique mode of binding revealed by cryo-EM studies. Further, we provide a comparative analysis of the mutational effects within previously emerged variant spikes and identify the structural role of mutations within the NTD and RBD in evading antibody neutralization. Our analysis shows that the highly mutated Gamma N-terminal domain exhibits considerable structural rearrangements, partially explaining its decreased neutralization by convalescent sera. Our results provide mechanistic insights into the structural, functional, and antigenic consequences of SARS-CoV-2 spike mutations and highlight a spike protein vulnerability that may be exploited to achieve broad protection against circulating variants. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27503.map.gz emd_27503.map.gz | 122.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27503-v30.xml emd-27503-v30.xml emd-27503.xml emd-27503.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27503_fsc.xml emd_27503_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_27503.png emd_27503.png | 58.1 KB | ||
| Filedesc metadata |  emd-27503.cif.gz emd-27503.cif.gz | 7 KB | ||
| Others |  emd_27503_half_map_1.map.gz emd_27503_half_map_1.map.gz emd_27503_half_map_2.map.gz emd_27503_half_map_2.map.gz | 226.5 MB 226.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27503 http://ftp.pdbj.org/pub/emdb/structures/EMD-27503 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27503 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27503 | HTTPS FTP |
-Validation report
| Summary document |  emd_27503_validation.pdf.gz emd_27503_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27503_full_validation.pdf.gz emd_27503_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_27503_validation.xml.gz emd_27503_validation.xml.gz | 19.3 KB | Display | |
| Data in CIF |  emd_27503_validation.cif.gz emd_27503_validation.cif.gz | 25.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27503 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27503 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27503 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27503 | HTTPS FTP |
-Related structure data
| Related structure data |  8dljMC  8dliC  8dlkC  8dllC  8dlmC  8dlnC  8dloC  8dlpC  8dlqC  8dlrC  8dlsC  8dltC  8dluC  8dlvC  8dlwC  8dlxC  8dlyC  8dlzC  8dm0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27503.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27503.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_27503_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
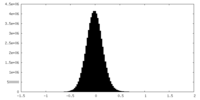
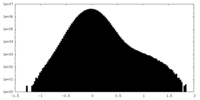
| Density Histograms |
-Half map: Half Map 2
| File | emd_27503_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
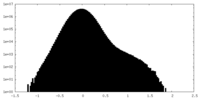
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2
| Entire | Name: SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2
| Supramolecule | Name: SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: SARS-CoV-2 Alpha (B.1.1.7) spike protein
| Supramolecule | Name: SARS-CoV-2 Alpha (B.1.1.7) spike protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: human ACE2
| Supramolecule | Name: human ACE2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 142.522609 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFQPTYGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIDDTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ GVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSHG SASSVASQSI IAYTMSLGAE NSVAYSNNSI AIPINFTI S VTTEILPVSM TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGF NFSQILPDPS KPSKRSPIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFG AGPALQIPFP MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STPSALGKLQ DVVNQNAQAL N TLVKQLSS NFGAISSVLN DILARLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RV DFCGKGY HLMSFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT HNT FVSGNC DVVIGIVNNT VYDPLQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDL QELGK YEQGSGYIPE APRDGQAYVR KDGEWVLLST FLGRSLEVLF QGPGHHHHHH HHSAWSHPQF EKGGGSGGGG SGGSA WSHP QFEK UniProtKB: Spike glycoprotein |
-Macromolecule #2: Processed angiotensin-converting enzyme 2
| Macromolecule | Name: Processed angiotensin-converting enzyme 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: angiotensin-converting enzyme 2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 70.386992 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSTIEEQAKT FLDKFNHEAE DLFYQSSLAS WNYNTNITEE NVQNMNNAGD KWSAFLKEQS TLAQMYPLQE IQNLTVKLQL QALQQNGSS VLSEDKSKRL NTILNTMSTI YSTGKVCNPD NPQECLLLEP GLNEIMANSL DYNERLWAWE SWRSEVGKQL R PLYEEYVV ...String: QSTIEEQAKT FLDKFNHEAE DLFYQSSLAS WNYNTNITEE NVQNMNNAGD KWSAFLKEQS TLAQMYPLQE IQNLTVKLQL QALQQNGSS VLSEDKSKRL NTILNTMSTI YSTGKVCNPD NPQECLLLEP GLNEIMANSL DYNERLWAWE SWRSEVGKQL R PLYEEYVV LKNEMARANH YEDYGDYWRG DYEVNGVDGY DYSRGQLIED VEHTFEEIKP LYEHLHAYVR AKLMNAYPSY IS PIGCLPA HLLGDMWGRF WTNLYSLTVP FGQKPNIDVT DAMVDQAWDA QRIFKEAEKF FVSVGLPNMT QGFWENSMLT DPG NVQKAV CHPTAWDLGK GDFRILMCTK VTMDDFLTAH HEMGHIQYDM AYAAQPFLLR NGANEGFHEA VGEIMSLSAA TPKH LKSIG LLSPDFQEDN ETEINFLLKQ ALTIVGTLPF TYMLEKWRWM VFKGEIPKDQ WMKKWWEMKR EIVGVVEPVP HDETY CDPA SLFHVSNDYS FIRYYTRTLY QFQFQEALCQ AAKHEGPLHK CDISNSTEAG QKLFNMLRLG KSEPWTLALE NVVGAK NMN VRPLLNYFEP LFTWLKDQNK NSFVGWSTDW SPYADHHHHH HHH UniProtKB: Angiotensin-converting enzyme 2 |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 28 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)