[English] 日本語
 Yorodumi
Yorodumi- EMDB-27499: Cryo-EM structure of human ferroportin/slc40 bound to Co2+ in nanodisc -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human ferroportin/slc40 bound to Co2+ in nanodisc | |||||||||||||||
 Map data Map data | Unsharpened map of human ferroportin/slc40 bound to Co2 in nanodisc | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | SLC40 / Fpn / ferroportin / iron / transporter / cobalt / human / nanodisc / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationspleen trabecula formation / iron ion export across plasma membrane / Defective SLC40A1 causes hemochromatosis 4 (HFE4) (duodenum) / Defective SLC40A1 causes hemochromatosis 4 (HFE4) (macrophages) / Defective CP causes aceruloplasminemia (ACERULOP) / Metal ion SLC transporters / lymphocyte homeostasis / iron ion transmembrane transporter activity / iron ion transmembrane transport / ferrous iron transmembrane transporter activity ...spleen trabecula formation / iron ion export across plasma membrane / Defective SLC40A1 causes hemochromatosis 4 (HFE4) (duodenum) / Defective SLC40A1 causes hemochromatosis 4 (HFE4) (macrophages) / Defective CP causes aceruloplasminemia (ACERULOP) / Metal ion SLC transporters / lymphocyte homeostasis / iron ion transmembrane transporter activity / iron ion transmembrane transport / ferrous iron transmembrane transporter activity / endothelium development / peptide hormone binding / establishment of localization in cell / Iron uptake and transport / multicellular organismal-level iron ion homeostasis / synaptic vesicle / basolateral plasma membrane / intracellular iron ion homeostasis / transcription by RNA polymerase II / negative regulation of apoptotic process / apoptotic process / positive regulation of transcription by RNA polymerase II / nucleoplasm / identical protein binding / membrane / metal ion binding / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||||||||
 Authors Authors | Shen J / Wilbon AS / Pan Y / Zhou M | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2023 Journal: PLoS Biol / Year: 2023Title: Structural basis of ferroportin inhibition by minihepcidin PR73. Authors: Azaan Saalim Wilbon / Jiemin Shen / Piotr Ruchala / Ming Zhou / Yaping Pan /  Abstract: Ferroportin (Fpn) is the only known iron exporter in humans and is essential for maintaining iron homeostasis. Fpn activity is suppressed by hepcidin, an endogenous peptide hormone, which inhibits ...Ferroportin (Fpn) is the only known iron exporter in humans and is essential for maintaining iron homeostasis. Fpn activity is suppressed by hepcidin, an endogenous peptide hormone, which inhibits iron export and promotes endocytosis of Fpn. Hepcidin deficiency leads to hemochromatosis and iron-loading anemia. Previous studies have shown that small peptides that mimic the first few residues of hepcidin, i.e., minihepcidins, are more potent than hepcidin. However, the mechanism of enhanced inhibition by minihepcidins remains unclear. Here, we report the structure of human ferroportin in complex with a minihepcidin, PR73 that mimics the first 9 residues of hepcidin, at 2.7 Å overall resolution. The structure reveals novel interactions that were not present between Fpn and hepcidin. We validate PR73-Fpn interactions through binding and transport assays. These results provide insights into how minihepcidins increase inhibition potency and will guide future development of Fpn inhibitors. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27499.map.gz emd_27499.map.gz | 15.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27499-v30.xml emd-27499-v30.xml emd-27499.xml emd-27499.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27499.png emd_27499.png | 79.6 KB | ||
| Filedesc metadata |  emd-27499.cif.gz emd-27499.cif.gz | 6.4 KB | ||
| Others |  emd_27499_half_map_1.map.gz emd_27499_half_map_1.map.gz emd_27499_half_map_2.map.gz emd_27499_half_map_2.map.gz | 28.3 MB 28.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27499 http://ftp.pdbj.org/pub/emdb/structures/EMD-27499 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27499 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27499 | HTTPS FTP |
-Validation report
| Summary document |  emd_27499_validation.pdf.gz emd_27499_validation.pdf.gz | 774.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27499_full_validation.pdf.gz emd_27499_full_validation.pdf.gz | 774.5 KB | Display | |
| Data in XML |  emd_27499_validation.xml.gz emd_27499_validation.xml.gz | 10.9 KB | Display | |
| Data in CIF |  emd_27499_validation.cif.gz emd_27499_validation.cif.gz | 12.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27499 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27499 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27499 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27499 | HTTPS FTP |
-Related structure data
| Related structure data |  8dl8MC  8dl7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27499.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27499.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map of human ferroportin/slc40 bound to Co2 in nanodisc | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
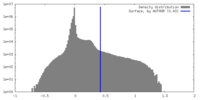
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half A map of human ferroportin/slc40 bound to Co2 in nanodisc
| File | emd_27499_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half_A map of human ferroportin/slc40 bound to Co2 in nanodisc | ||||||||||||
| Projections & Slices |
| ||||||||||||
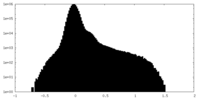
| Density Histograms |
-Half map: Half B map of human ferroportin/slc40 bound to Co2 in nanodisc
| File | emd_27499_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half_B map of human ferroportin/slc40 bound to Co2 in nanodisc | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of human ferroportin/slc40 bound to Co2+ in nanodisc
| Entire | Name: Cryo-EM structure of human ferroportin/slc40 bound to Co2+ in nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of human ferroportin/slc40 bound to Co2+ in nanodisc
| Supramolecule | Name: Cryo-EM structure of human ferroportin/slc40 bound to Co2+ in nanodisc type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 40 member 1
| Macromolecule | Name: Solute carrier family 40 member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 63.386621 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTRAGDHNRQ RGCCGSLADY LTSAKFLLYL GHSLSTWGDR MWHFAVSVFL VELYGNSLLL TAVYGLVVAG SVLVLGAIIG DWVDKNARL KVAQTSLVVQ NVSVILCGII LMMVFLHKHE LLTMYHGWVL TSCYILIITI ANIANLASTA TAITIQRDWI V VVAGEDRS ...String: MTRAGDHNRQ RGCCGSLADY LTSAKFLLYL GHSLSTWGDR MWHFAVSVFL VELYGNSLLL TAVYGLVVAG SVLVLGAIIG DWVDKNARL KVAQTSLVVQ NVSVILCGII LMMVFLHKHE LLTMYHGWVL TSCYILIITI ANIANLASTA TAITIQRDWI V VVAGEDRS KLANMNATIR RIDQLTNILA PMAVGQIMTF GSPVIGCGFI SGWNLVSMCV EYVLLWKVYQ KTPALAVKAG LK EEETELK QLNLHKDTEP KPLEGTHLMG VKDSNIHELE HEQEPTCASQ MAEPFRTFRD GWVSYYNQPV FLAGMGLAFL YMT VLGFDC ITTGYAYTQG LSGSILSILM GASAITGIMG TVAFTWLRRK CGLVRTGLIS GLAQLSCLIL CVISVFMPGS PLDL SVSPF EDIRSRFIQG ESITPTKIPE ITTEIYMSNG SNSANIVPET SPESVPIISV SLLFAGVIAA RIGLWSFDLT VTQLL QENV IESERGIING VQNSMNYLLD LLHFIMVILA PNPEAFGLLV LISVSFVAMG HIMYFRFAQN TLGNKLFACG PDAKEV RKE NQANTSVVEN LYFQ UniProtKB: Ferroportin |
-Macromolecule #2: 11F9 light-chain
| Macromolecule | Name: 11F9 light-chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.493885 KDa |
| Sequence | String: DIVMTQSQKF MSTSVGDRVS ITCKASQNVG TAVAWYQKKP GQSPKLLIYS ASNRYSGVPD RFTGSGSGTD FTLTISNMQS EDLADYFCQ QYGSYPLTFG SGTKLEIKEA EAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS ...String: DIVMTQSQKF MSTSVGDRVS ITCKASQNVG TAVAWYQKKP GQSPKLLIYS ASNRYSGVPD RFTGSGSGTD FTLTISNMQS EDLADYFCQ QYGSYPLTFG SGTKLEIKEA EAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS KDSTYSMSST LTLTKDEYER HNSYTCEATH KTSTSPIVKS FNRNE |
-Macromolecule #3: 11F9 heavy-chain
| Macromolecule | Name: 11F9 heavy-chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.815039 KDa |
| Sequence | String: MKCSWVIFFL MAVVTGVNSE VQLQQSGAEL VRPGALVKLS CKASGFNIKD YYMHWVKERP EQGLEWIGWI DPENGNTIYD PKFQGKASI TADTSSNTAY LQLSSLTSED TAVYYCARKR GYYGPYFDYW GQGTTLTVSS KTTAPSVYPL APVCGDTTGS S VTLGCLVK ...String: MKCSWVIFFL MAVVTGVNSE VQLQQSGAEL VRPGALVKLS CKASGFNIKD YYMHWVKERP EQGLEWIGWI DPENGNTIYD PKFQGKASI TADTSSNTAY LQLSSLTSED TAVYYCARKR GYYGPYFDYW GQGTTLTVSS KTTAPSVYPL APVCGDTTGS S VTLGCLVK GYFPEPVTLT WNSGSLSSGV HTFPAVLQSG LYTLSSSVTV TSSTWPSQSI TCNVAHPASS TKVDKKIEPA |
-Macromolecule #4: COBALT (II) ION
| Macromolecule | Name: COBALT (II) ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: CO |
|---|---|
| Molecular weight | Theoretical: 58.933 Da |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 305 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.25 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 215164 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)