+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
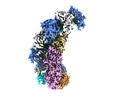
| Title | type I-C Cascade bound to AcrIC4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / type I-C / anti-CRISPR / Cascade / type I-C Cascade / AcrIC4 / DNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / defense response to virus / endonuclease activity / Hydrolases; Acting on ester bonds / RNA binding Similarity search - Function | |||||||||
| Biological species |  Desulfovibrio vulgaris (bacteria) / Desulfovibrio vulgaris (bacteria) /  Pseudomonas aeruginosa phage DMS3mVir (virus) Pseudomonas aeruginosa phage DMS3mVir (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | O'Brien RE / Bravo JPK / Ramos D / Hibshman GN / Wright JT / Taylor DW | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Structural snapshots of R-loop formation by a type I-C CRISPR Cascade. Authors: Roisin E O'Brien / Jack P K Bravo / Delisa Ramos / Grace N Hibshman / Jacquelyn T Wright / David W Taylor /  Abstract: Type I CRISPR-Cas systems employ multi-subunit Cascade effector complexes to target foreign nucleic acids for destruction. Here, we present structures of D. vulgaris type I-C Cascade at various ...Type I CRISPR-Cas systems employ multi-subunit Cascade effector complexes to target foreign nucleic acids for destruction. Here, we present structures of D. vulgaris type I-C Cascade at various stages of double-stranded (ds)DNA target capture, revealing mechanisms that underpin PAM recognition and Cascade allosteric activation. We uncover an interesting mechanism of non-target strand (NTS) DNA stabilization via stacking interactions with the "belly" subunits, securing the NTS in place. This "molecular seatbelt" mechanism facilitates efficient R-loop formation and prevents dsDNA reannealing. Additionally, we provide structural insights into how two anti-CRISPR (Acr) proteins utilize distinct strategies to achieve a shared mechanism of type I-C Cascade inhibition by blocking PAM scanning. These observations form a structural basis for directional R-loop formation and reveal how different Acr proteins have converged upon common molecular mechanisms to efficiently shut down CRISPR immunity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27409.map.gz emd_27409.map.gz | 267 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27409-v30.xml emd-27409-v30.xml emd-27409.xml emd-27409.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27409.png emd_27409.png | 65.9 KB | ||
| Filedesc metadata |  emd-27409.cif.gz emd-27409.cif.gz | 6.2 KB | ||
| Others |  emd_27409_half_map_1.map.gz emd_27409_half_map_1.map.gz emd_27409_half_map_2.map.gz emd_27409_half_map_2.map.gz | 262.4 MB 262.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27409 http://ftp.pdbj.org/pub/emdb/structures/EMD-27409 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27409 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27409 | HTTPS FTP |
-Validation report
| Summary document |  emd_27409_validation.pdf.gz emd_27409_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27409_full_validation.pdf.gz emd_27409_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_27409_validation.xml.gz emd_27409_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  emd_27409_validation.cif.gz emd_27409_validation.cif.gz | 19.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27409 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27409 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27409 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27409 | HTTPS FTP |
-Related structure data
| Related structure data |  8dfoMC  8dejC  8dexC  8dfaC  8dfsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27409.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27409.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.94 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27409_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_27409_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : type I-C Cascade bound to AcrIC4
| Entire | Name: type I-C Cascade bound to AcrIC4 |
|---|---|
| Components |
|
-Supramolecule #1: type I-C Cascade bound to AcrIC4
| Supramolecule | Name: type I-C Cascade bound to AcrIC4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|
-Macromolecule #1: pre-crRNA processing endonuclease
| Macromolecule | Name: pre-crRNA processing endonuclease / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Desulfovibrio vulgaris (bacteria) Desulfovibrio vulgaris (bacteria)Strain: ATCC 29579 / DSM 644 / NCIMB 8303 / VKM B-1760 / Hildenborough |
| Molecular weight | Theoretical: 25.977857 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTHGAVKTYG IRLRVWGDYA CFTRPEMKVE RVSYDVMPPS AARGILEAIH WKPAIRWIVD RIHVLRPIVF DNVRRNEVSS KIPKPNPAT AMRDRKPLYF LVDDGSNRQQ RAATLLRNVD YVIEAHFELT DKAGAEDNAG KHLDIFRRRA RAGQSFQQPC L GCREFPAS ...String: MTHGAVKTYG IRLRVWGDYA CFTRPEMKVE RVSYDVMPPS AARGILEAIH WKPAIRWIVD RIHVLRPIVF DNVRRNEVSS KIPKPNPAT AMRDRKPLYF LVDDGSNRQQ RAATLLRNVD YVIEAHFELT DKAGAEDNAG KHLDIFRRRA RAGQSFQQPC L GCREFPAS FELLEGDVPL SCYAGEKRDL GYMLLDIDFE RDMTPLFFKA VMEDGVITPP SRTSPEVRA UniProtKB: pre-crRNA processing endonuclease |
-Macromolecule #2: CRISPR-associated protein, TM1801 family
| Macromolecule | Name: CRISPR-associated protein, TM1801 family / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfovibrio vulgaris (bacteria) Desulfovibrio vulgaris (bacteria)Strain: ATCC 29579 / DSM 644 / NCIMB 8303 / VKM B-1760 / Hildenborough |
| Molecular weight | Theoretical: 32.358912 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTAIANRYEF VLLFDVENGN PNGDPDAGNM PRIDPETGHG LVTDVCLKRK IRNHVALTKE GAERFNIYIQ EKAILNETHE RAYTACDLK PEPKKLPKKV EDAKRVTDWM CTNFYDIRTF GAVMTTEVNC GQVRGPVQMA FARSVEPVVP QEVSITRMAV T TKAEAEKQ ...String: MTAIANRYEF VLLFDVENGN PNGDPDAGNM PRIDPETGHG LVTDVCLKRK IRNHVALTKE GAERFNIYIQ EKAILNETHE RAYTACDLK PEPKKLPKKV EDAKRVTDWM CTNFYDIRTF GAVMTTEVNC GQVRGPVQMA FARSVEPVVP QEVSITRMAV T TKAEAEKQ QGDNRTMGRK HIVPYGLYVA HGFISAPLAE KTGFSDEDLT LFWDALVNMF EHDRSAARGL MSSRKLIVFK HQ NRLGNAP AHKLFDLVKV SRAEGSSGPA RSFADYAVTV GQAPEGVEVK EML UniProtKB: CRISPR-associated protein, TM1801 family |
-Macromolecule #3: CRISPR-associated protein, CT1133 family
| Macromolecule | Name: CRISPR-associated protein, CT1133 family / type: protein_or_peptide / ID: 3 / Details: Cas8c / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfovibrio vulgaris (bacteria) Desulfovibrio vulgaris (bacteria)Strain: ATCC 29579 / DSM 644 / NCIMB 8303 / VKM B-1760 / Hildenborough |
| Molecular weight | Theoretical: 68.123219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MILQALHGYY QRMSADPDAG MPPYGTSMEN ISFALVLDAK GTLRGIEDLR EQEGKKLRPR KMLVPIAEKK GNGIKPNFLW ENTSYILGV DAKGKQERTD KCHAAFIAHI KAYCDTADQD LAAVLQFLEH GEKDLSAFPV SEEVIGSNIV FRIEGEPGFV H ERPAARQA ...String: MILQALHGYY QRMSADPDAG MPPYGTSMEN ISFALVLDAK GTLRGIEDLR EQEGKKLRPR KMLVPIAEKK GNGIKPNFLW ENTSYILGV DAKGKQERTD KCHAAFIAHI KAYCDTADQD LAAVLQFLEH GEKDLSAFPV SEEVIGSNIV FRIEGEPGFV H ERPAARQA WANCLNRREQ GLCGQCLITG ERQKPIAQLH PSIKGGRDGV RGAQAVASIV SFNNTAFESY GKEQSINAPV SQ EAAFSYV TALNYLLNPS NRQKVTIADA TVVFWAERSS PAEDIFAGMF DPPSTTAKPE SSNGTPPEDS EEGSQPDTAR DDP HAAARM HDLLVAIRSG KRATDIMPDM DESVRFHVLG LSPNAARLSV RFWEVDTVGH MLDKVGRHYR ELEIIPQFNN EQEF PSLST LLRQTAVLNK TENISPVLAG GLFRAMLTGG PYPQSLLPAV LGRIRAEHAR PEDKSRYRLE VVTYYRAALI KAYLI RNRK LEVPVSLDPA RTDRPYLLGR LFAVLEKAQE DAVPGANATI KDRYLASASA NPGQVFHMLL KNASNHTAKL RKDPER KGS AIHYEIMMQE IIDNISDFPV TMSSDEQGLF MIGYYHQRKA LFTKKNKEN UniProtKB: CRISPR-associated protein, CT1133 family |
-Macromolecule #4: CRISPR-associated protein, CT1133 family
| Macromolecule | Name: CRISPR-associated protein, CT1133 family / type: protein_or_peptide / ID: 4 / Details: Cas11c / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfovibrio vulgaris (bacteria) Desulfovibrio vulgaris (bacteria)Strain: ATCC 29579 / DSM 644 / NCIMB 8303 / VKM B-1760 / Hildenborough |
| Molecular weight | Theoretical: 14.017981 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VSLDPARTDR PYLLGRLFAV LEKAQEDAVP GANATIKDRY LASASANPGQ VFHMLLKNAS NHTAKLRKDP ERKGSAIHYE IMMQEIIDN ISDFPVTMSS DEQGLFMIGY YHQRKALFTK KNKEN UniProtKB: CRISPR-associated protein, CT1133 family |
-Macromolecule #6: AcrIC4
| Macromolecule | Name: AcrIC4 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa phage DMS3mVir (virus) Pseudomonas aeruginosa phage DMS3mVir (virus) |
| Molecular weight | Theoretical: 6.659252 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDNKITPADE EKIREWLNCE EASVDNDGDV WVAVPMTGHW LSDEQKAKYI EWRGDET |
-Macromolecule #5: RNA (45-MER)
| Macromolecule | Name: RNA (45-MER) / type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Desulfovibrio vulgaris (bacteria) / Strain: Hildenborough Desulfovibrio vulgaris (bacteria) / Strain: Hildenborough |
| Molecular weight | Theoretical: 14.472646 KDa |
| Sequence | String: GGAUUGAAAC GCCAUGCUCA GGCUGGCGAG UGCGCGCCAC UCAUC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 21651 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)