[English] 日本語
 Yorodumi
Yorodumi- EMDB-27237: Cereblon~DDB1 in the Apo form with DDB1 in the hinged conformation -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cereblon~DDB1 in the Apo form with DDB1 in the hinged conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ubiquitin / CRL4 / Adaptor / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of monoatomic ion transmembrane transport / positive regulation by virus of viral protein levels in host cell / epigenetic programming in the zygotic pronuclei / spindle assembly involved in female meiosis / Cul4-RING E3 ubiquitin ligase complex / UV-damage excision repair / biological process involved in interaction with symbiont / regulation of mitotic cell cycle phase transition / WD40-repeat domain binding / Cul4A-RING E3 ubiquitin ligase complex ...negative regulation of monoatomic ion transmembrane transport / positive regulation by virus of viral protein levels in host cell / epigenetic programming in the zygotic pronuclei / spindle assembly involved in female meiosis / Cul4-RING E3 ubiquitin ligase complex / UV-damage excision repair / biological process involved in interaction with symbiont / regulation of mitotic cell cycle phase transition / WD40-repeat domain binding / Cul4A-RING E3 ubiquitin ligase complex / Cul4B-RING E3 ubiquitin ligase complex / ubiquitin ligase complex scaffold activity / negative regulation of reproductive process / negative regulation of developmental process / locomotory exploration behavior / cullin family protein binding / viral release from host cell / positive regulation of Wnt signaling pathway / ectopic germ cell programmed cell death / negative regulation of protein-containing complex assembly / positive regulation of viral genome replication / proteasomal protein catabolic process / positive regulation of gluconeogenesis / nucleotide-excision repair / positive regulation of protein-containing complex assembly / Recognition of DNA damage by PCNA-containing replication complex / DNA Damage Recognition in GG-NER / regulation of circadian rhythm / Wnt signaling pathway / Dual Incision in GG-NER / Transcription-Coupled Nucleotide Excision Repair (TC-NER) / Formation of TC-NER Pre-Incision Complex / Formation of Incision Complex in GG-NER / Dual incision in TC-NER / positive regulation of protein catabolic process / Gap-filling DNA repair synthesis and ligation in TC-NER / cellular response to UV / rhythmic process / site of double-strand break / Neddylation / ubiquitin-dependent protein catabolic process / protein-macromolecule adaptor activity / proteasome-mediated ubiquitin-dependent protein catabolic process / transmembrane transporter binding / Potential therapeutics for SARS / damaged DNA binding / chromosome, telomeric region / protein ubiquitination / DNA repair / apoptotic process / DNA damage response / protein-containing complex binding / negative regulation of apoptotic process / nucleolus / perinuclear region of cytoplasm / protein-containing complex / DNA binding / extracellular space / extracellular exosome / nucleoplasm / nucleus / metal ion binding / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
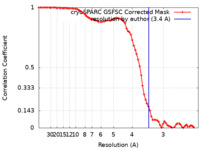
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Watson ER / Lander GC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Molecular glue CELMoD compounds are regulators of cereblon conformation. Authors: Edmond R Watson / Scott Novick / Mary E Matyskiela / Philip P Chamberlain / Andres H de la Peña / Jinyi Zhu / Eileen Tran / Patrick R Griffin / Ingrid E Wertz / Gabriel C Lander /  Abstract: Cereblon (CRBN) is a ubiquitin ligase (E3) substrate receptor protein co-opted by CRBN E3 ligase modulatory drug (CELMoD) agents that target therapeutically relevant proteins for degradation. Prior ...Cereblon (CRBN) is a ubiquitin ligase (E3) substrate receptor protein co-opted by CRBN E3 ligase modulatory drug (CELMoD) agents that target therapeutically relevant proteins for degradation. Prior crystallographic studies defined the drug-binding site within CRBN's thalidomide-binding domain (TBD), but the allostery of drug-induced neosubstrate binding remains unclear. We performed cryo-electron microscopy analyses of the DNA damage-binding protein 1 (DDB1)-CRBN apo complex and compared these structures with DDB1-CRBN in the presence of CELMoD compounds alone and complexed with neosubstrates. Association of CELMoD compounds to the TBD is necessary and sufficient for triggering CRBN allosteric rearrangement from an open conformation to the canonical closed conformation. The neosubstrate Ikaros only stably associates with the closed CRBN conformation, illustrating the importance of allostery for CELMoD compound efficacy and informing structure-guided design strategies to improve therapeutic efficacy. #1:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: Molecular glue CELMoD compounds are allosteric regulators of cereblon conformation Authors: Watson ER / Novick SJ / Matyskiela ME / Chamberlain PP / de la Pena AH / Zhu JY / Tran E / Griffin PR / Wertz IE / Lander GC | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27237.map.gz emd_27237.map.gz | 22.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27237-v30.xml emd-27237-v30.xml emd-27237.xml emd-27237.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27237_fsc.xml emd_27237_fsc.xml | 7.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_27237.png emd_27237.png | 107.2 KB | ||
| Filedesc metadata |  emd-27237.cif.gz emd-27237.cif.gz | 6.7 KB | ||
| Others |  emd_27237_additional_1.map.gz emd_27237_additional_1.map.gz emd_27237_half_map_1.map.gz emd_27237_half_map_1.map.gz emd_27237_half_map_2.map.gz emd_27237_half_map_2.map.gz | 21.6 MB 39.8 MB 39.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27237 http://ftp.pdbj.org/pub/emdb/structures/EMD-27237 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27237 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27237 | HTTPS FTP |
-Validation report
| Summary document |  emd_27237_validation.pdf.gz emd_27237_validation.pdf.gz | 897.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27237_full_validation.pdf.gz emd_27237_full_validation.pdf.gz | 897.1 KB | Display | |
| Data in XML |  emd_27237_validation.xml.gz emd_27237_validation.xml.gz | 15 KB | Display | |
| Data in CIF |  emd_27237_validation.cif.gz emd_27237_validation.cif.gz | 19.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27237 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27237 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27237 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27237 | HTTPS FTP |
-Related structure data
| Related structure data |  8d7xMC  8cvpC  8d7uC  8d7vC  8d7wC  8d7yC  8d7zC  8d80C  8d81C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27237.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27237.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_27237_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_27237_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_27237_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cereblon~DDB1 in the Apo form with DDB1 in the hinged conformation
| Entire | Name: Cereblon~DDB1 in the Apo form with DDB1 in the hinged conformation |
|---|---|
| Components |
|
-Supramolecule #1: Cereblon~DDB1 in the Apo form with DDB1 in the hinged conformation
| Supramolecule | Name: Cereblon~DDB1 in the Apo form with DDB1 in the hinged conformation type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 177 KDa |
-Macromolecule #1: DNA damage-binding protein 1
| Macromolecule | Name: DNA damage-binding protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 127.097469 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSYNYVVTAQ KPTAVNGCVT GHFTSAEDLN LLIAKNTRLE IYVVTAEGLR PVKEVGMYGK IAVMELFRPK GESKDLLFIL TAKYNACIL EYKQSGESID IITRAHGNVQ DRIGRPSETG IIGIIDPECR MIGLRLYDGL FKVIPLDRDN KELKAFNIRL E ELHVIDVK ...String: MSYNYVVTAQ KPTAVNGCVT GHFTSAEDLN LLIAKNTRLE IYVVTAEGLR PVKEVGMYGK IAVMELFRPK GESKDLLFIL TAKYNACIL EYKQSGESID IITRAHGNVQ DRIGRPSETG IIGIIDPECR MIGLRLYDGL FKVIPLDRDN KELKAFNIRL E ELHVIDVK FLYGCQAPTI CFVYQDPQGR HVKTYEVSLR EKEFNKGPWK QENVEAEASM VIAVPEPFGG AIIIGQESIT YH NGDKYLA IAPPIIKQST IVCHNRVDPN GSRYLLGDME GRLFMLLLEK EEQMDGTVTL KDLRVELLGE TSIAECLTYL DNG VVFVGS RLGDSQLVKL NVDSNEQGSY VVAMETFTNL GPIVDMCVVD LERQGQGQLV TCSGAFKEGS LRIIRNGIGI HEHA SIDLP GIKGLWPLRS DPNRETDDTL VLSFVGQTRV LMLNGEEVEE TELMGFVDDQ QTFFCGNVAH QQLIQITSAS VRLVS QEPK ALVSEWKEPQ AKNISVASCN SSQVVVAVGR ALYYLQIHPQ ELRQISHTEM EHEVACLDIT PLGDSNGLSP LCAIGL WTD ISARILKLPS FELLHKEMLG GEIIPRSILM TTFESSHYLL CALGDGALFY FGLNIETGLL SDRKKVTLGT QPTVLRT FR SLSTTNVFAC SDRPTVIYSS NHKLVFSNVN LKEVNYMCPL NSDGYPDSLA LANNSTLTIG TIDEIQKLHI RTVPLYES P RKICYQEVSQ CFGVLSSRIE VQDTSGGTTA LRPSASTQAL SSSVSSSKLF SSSTAPHETS FGEEVEVHNL LIIDQHTFE VLHAHQFLQN EYALSLVSCK LGKDPNTYFI VGTAMVYPEE AEPKQGRIVV FQYSDGKLQT VAEKEVKGAV YSMVEFNGKL LASINSTVR LYEWTTEKEL RTECNHYNNI MALYLKTKGD FILVGDLMRS VLLLAYKPME GNFEEIARDF NPNWMSAVEI L DDDNFLGA ENAFNLFVCQ KDSAATTDEE RQHLQEVGLF HLGEFVNVFC HGSLVMQNLG ETSTPTQGSV LFGTVNGMIG LV TSLSESW YNLLLDMQNR LNKVIKSVGK IEHSFWRSFH TERKTEPATG FIDGDLIESF LDISRPKMQE VVANLQYDDG SGM KREATA DDLIKVVEEL TRIH UniProtKB: DNA damage-binding protein 1 |
-Macromolecule #2: Protein cereblon
| Macromolecule | Name: Protein cereblon / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.603676 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGEGDQQDA AHNMGNHLPL LPAESEEEDE MEVEDQDSKE AKKPNIINFD TSLPTSHTYL GADMEEFHGR TLHDDDSCQV IPVLPQVMM ILIPGQTLPL QLFHPQEVSM VRNLIQKDRT FAVLAYSNVQ EREAQFGTTA EIYAYREEQD FGIEIVKVKA I GRQRFKVL ...String: MAGEGDQQDA AHNMGNHLPL LPAESEEEDE MEVEDQDSKE AKKPNIINFD TSLPTSHTYL GADMEEFHGR TLHDDDSCQV IPVLPQVMM ILIPGQTLPL QLFHPQEVSM VRNLIQKDRT FAVLAYSNVQ EREAQFGTTA EIYAYREEQD FGIEIVKVKA I GRQRFKVL ELRTQSDGIQ QAKVQILPEC VLPSTMSAVQ LESLNKCQIF PSKPVSREDQ CSYKWWQKYQ KRKFHCANLT SW PRWLYSL YDAETLMDRI KKQLREWDEN LKDDSLPSNP IDFSYRVAAC LPIDDVLRIQ LLKIGSAIQR LRCELDIMNK CTS LCCKQC QETEITTKNE IFSLSLCGPM AAYVNPHGYV HETLTVYKAC NLNLIGRPST EHSWFPGYAW TVAQCKICAS HIGW KFTAT KKDMSPQKFW GLTRSALLPT IPDTEDEISP DKVILCL UniProtKB: Protein cereblon |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 20 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: 20mM HEPES pH 7, 240mM NaCl, 3mM TCEP |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER / Details: Manual plunge in 4 degree C cold room. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 62.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)