+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
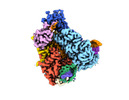
| Title | Structure of Cas12a2 ternary complex | ||||||||||||
 Map data Map data | Unsharpened map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Cas12a2 / CRISPR / Nuclease / DNA Binding Protein-RNA-DNA complex | ||||||||||||
| Function / homology | Transposase IS605, OrfB, C-terminal / Putative transposase DNA-binding domain / DNA binding / Cas12f1-like TNB domain-containing protein Function and homology information Function and homology information | ||||||||||||
| Biological species |  Sulfuricurvum sp. PC08-66 (bacteria) / synthetic construct (others) Sulfuricurvum sp. PC08-66 (bacteria) / synthetic construct (others) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.74 Å | ||||||||||||
 Authors Authors | Bravo JPK / Taylor DW | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: RNA targeting unleashes indiscriminate nuclease activity of CRISPR-Cas12a2. Authors: Jack P K Bravo / Thomson Hallmark / Bronson Naegle / Chase L Beisel / Ryan N Jackson / David W Taylor /   Abstract: Cas12a2 is a CRISPR-associated nuclease that performs RNA-guided, sequence-nonspecific degradation of single-stranded RNA, single-stranded DNA and double-stranded DNA following recognition of a ...Cas12a2 is a CRISPR-associated nuclease that performs RNA-guided, sequence-nonspecific degradation of single-stranded RNA, single-stranded DNA and double-stranded DNA following recognition of a complementary RNA target, culminating in abortive infection. Here we report structures of Cas12a2 in binary, ternary and quaternary complexes to reveal a complete activation pathway. Our structures reveal that Cas12a2 is autoinhibited until binding a cognate RNA target, which exposes the RuvC active site within a large, positively charged cleft. Double-stranded DNA substrates are captured through duplex distortion and local melting, stabilized by pairs of 'aromatic clamp' residues that are crucial for double-stranded DNA degradation and in vivo immune system function. Our work provides a structural basis for this mechanism of abortive infection to achieve population-level immunity, which can be leveraged to create rational mutants that degrade a spectrum of collateral substrates. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27179.map.gz emd_27179.map.gz | 108.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27179-v30.xml emd-27179-v30.xml emd-27179.xml emd-27179.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27179.png emd_27179.png | 72.5 KB | ||
| Filedesc metadata |  emd-27179.cif.gz emd-27179.cif.gz | 6.6 KB | ||
| Others |  emd_27179_additional_1.map.gz emd_27179_additional_1.map.gz emd_27179_half_map_1.map.gz emd_27179_half_map_1.map.gz emd_27179_half_map_2.map.gz emd_27179_half_map_2.map.gz | 203.9 MB 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27179 http://ftp.pdbj.org/pub/emdb/structures/EMD-27179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27179 | HTTPS FTP |
-Validation report
| Summary document |  emd_27179_validation.pdf.gz emd_27179_validation.pdf.gz | 900.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27179_full_validation.pdf.gz emd_27179_full_validation.pdf.gz | 900 KB | Display | |
| Data in XML |  emd_27179_validation.xml.gz emd_27179_validation.xml.gz | 15.7 KB | Display | |
| Data in CIF |  emd_27179_validation.cif.gz emd_27179_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27179 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27179 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27179 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27179 | HTTPS FTP |
-Related structure data
| Related structure data |  8d4aMC  8d49C  8d4bC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27179.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27179.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.94 Å | ||||||||||||||||||||||||||||||||||||
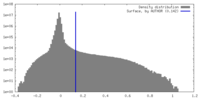
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
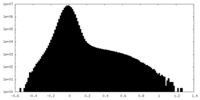
-Additional map: Additional Map
| File | emd_27179_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
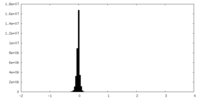
| Density Histograms |
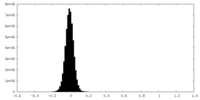
-Half map: Half Map 1
| File | emd_27179_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
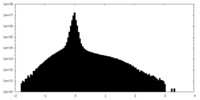
| Density Histograms |
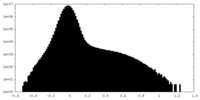
-Half map: Half Map 2
| File | emd_27179_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
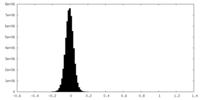
| Density Histograms |
- Sample components
Sample components
-Entire : Cas12a2 ternary complex
| Entire | Name: Cas12a2 ternary complex |
|---|---|
| Components |
|
-Supramolecule #1: Cas12a2 ternary complex
| Supramolecule | Name: Cas12a2 ternary complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Sulfuricurvum sp. PC08-66 (bacteria) Sulfuricurvum sp. PC08-66 (bacteria) |
-Macromolecule #1: OrfB_Zn_ribbon domain-containing protein
| Macromolecule | Name: OrfB_Zn_ribbon domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sulfuricurvum sp. PC08-66 (bacteria) Sulfuricurvum sp. PC08-66 (bacteria) |
| Molecular weight | Theoretical: 143.172797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLHAFTNQYQ LSKTLRFGAT LKEDEKKCKS HEELKGFVDI SYENMKSSAT IAESLNENEL VKKCERCYSE IVKFHNAWEK IYYRTDQIA VYKDFYRQLS RKARFDAGKQ NSQLITLASL CGMYQGAKLS RYITNYWKDN ITRQKSFLKD FSQQLHQYTR A LEKSDKAH ...String: MLHAFTNQYQ LSKTLRFGAT LKEDEKKCKS HEELKGFVDI SYENMKSSAT IAESLNENEL VKKCERCYSE IVKFHNAWEK IYYRTDQIA VYKDFYRQLS RKARFDAGKQ NSQLITLASL CGMYQGAKLS RYITNYWKDN ITRQKSFLKD FSQQLHQYTR A LEKSDKAH TKPNLINFNK TFMVLANLVN EIVIPLSNGA ISFPNISKLE DGEESHLIEF ALNDYSQLSE LIGELKDAIA TN GGYTPFA KVTLNHYTAE QKPHVFKNDI DAKIRELKLI GLVETLKGKS SEQIEEYFSN LDKFSTYNDR NQSVIVRTQC FKY KPIPFL VKHQLAKYIS EPNGWDEDAV AKVLDAVGAI RSPAHDYANN QEGFDLNHYP IKVAFDYAWE QLANSLYTTV TFPQ EMCEK YLNSIYGCEV SKEPVFKFYA DLLYIRKNLA VLEHKNNLPS NQEEFICKIN NTFENIVLPY KISQFETYKK DILAW INDG HDHKKYTDAK QQLGFIRGGL KGRIKAEEVS QKDKYGKIKS YYENPYTKLT NEFKQISSTY GKTFAELRDK FKEKNE ITK ITHFGIIIED KNRDRYLLAS ELKHEQINHV STILNKLDKS SEFITYQVKS LTSKTLIKLI KNHTTKKGAI SPYADFH TS KTGFNKNEIE KNWDNYKREQ VLVEYVKDCL TDSTMAKNQN WAEFGWNFEK CNSYEDIEHE IDQKSYLLQS DTISKQSI A SLVEGGCLLL PIINQDITSK ERKDKNQFSK DWNHIFEGSK EFRLHPEFAV SYRTPIEGYP VQKRYGRLQF VCAFNAHIV PQNGEFINLK KQIENFNDED VQKRNVTEFN KKVNHALSDK EYVVIGIDRG LKQLATLCVL DKRGKILGDF EIYKKEFVRA EKRSESHWE HTQAETRHIL DLSNLRVETT IEGKKVLVDQ SLTLVKKNRD TPDEEATEEN KQKIKLKQLS YIRKLQHKMQ T NEQDVLDL INNEPSDEEF KKRIEGLISS FGEGQKYADL PINTMREMIS DLQGVIARGN NQTEKNKIIE LDAADNLKQG IV ANMIGIV NYIFAKYSYK AYISLEDLSR AYGGAKSGYD GRYLPSTSQD EDVDFKEQQN QMLAGLGTYQ FFEMQLLKKL QKI QSDNTV LRFVPAFRSA DNYRNILRLE ETKYKSKPFG VVHFIDPKFT SKKCPVCSKT NVYRDKDDIL VCKECGFRSD SQLK ERENN IHYIHNGDDN GAYHIALKSV ENLIQMK UniProtKB: Cas12f1-like TNB domain-containing protein |
-Macromolecule #2: RNA (41-MER)
| Macromolecule | Name: RNA (41-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 13.151831 KDa |
| Sequence | String: AUUUCUACUA UUGUAGAUUG GAGCAACACC UGAAGAAGGC U |
-Macromolecule #3: RNA (28-MER)
| Macromolecule | Name: RNA (28-MER) / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.934285 KDa |
| Sequence | String: AGCCUUCUUC AGGUGUUGCU UUAGAAAG |
-Macromolecule #4: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3') / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 3.301163 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT) |
-Macromolecule #5: DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3')
| Macromolecule | Name: DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 3.400317 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA) |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.74 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 104857 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)