[English] 日本語
 Yorodumi
Yorodumi- EMDB-27145: Structure of Acidothermus cellulolyticus Cas9 ternary complex (Ta... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Target bound) | |||||||||
 Map data Map data | Accas9 ternary complex (Target bound) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cas9 / AceCas9 / Crispr / Target bound / RNA BINDING PROTEIN / RNA BINDING PROTEIN-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationdefense response to virus / endonuclease activity / DNA binding / RNA binding / zinc ion binding Similarity search - Function | |||||||||
| Biological species |  Acidothermus cellulolyticus 11B (bacteria) / synthetic construct (others) Acidothermus cellulolyticus 11B (bacteria) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.78 Å | |||||||||
 Authors Authors | Rai J / Das A / Li H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Catal / Year: 2023 Journal: Nat Catal / Year: 2023Title: Coupled catalytic states and the role of metal coordination in Cas9. Authors: Anuska Das / Jay Rai / Mitchell O Roth / Yuerong Shu / Megan L Medina / Mackenzie R Barakat / Hong Li /  Abstract: Controlling the activity of the CRISPR-Cas9 system is essential to its safe adoption for clinical and research applications. Although the conformational dynamics of Cas9 are known to control its ...Controlling the activity of the CRISPR-Cas9 system is essential to its safe adoption for clinical and research applications. Although the conformational dynamics of Cas9 are known to control its enzymatic activity, details of how Cas9 influences the catalytic processes at both nuclease domains remain elusive. Here we report five cryo-electron microscopy structures of the active Cas9 complex along the reaction path at 2.2-2.9 Å resolution. We observed that a large movement in one nuclease domain, triggered by the cognate DNA, results in noticeable changes in the active site of the other domain that is required for metal coordination and catalysis. Furthermore, the conformations synchronize the reaction intermediates, enabling coupled cutting of the two DNA strands. Consistent with the roles of conformations in organizing the active sites, adjustments to the metal-coordination residues lead to altered metal specificity of Cas9 and commonly used Cas9 in cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27145.map.gz emd_27145.map.gz | 100.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27145-v30.xml emd-27145-v30.xml emd-27145.xml emd-27145.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27145.png emd_27145.png | 73.8 KB | ||
| Filedesc metadata |  emd-27145.cif.gz emd-27145.cif.gz | 6.8 KB | ||
| Others |  emd_27145_half_map_1.map.gz emd_27145_half_map_1.map.gz emd_27145_half_map_2.map.gz emd_27145_half_map_2.map.gz | 84.2 MB 84.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27145 http://ftp.pdbj.org/pub/emdb/structures/EMD-27145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27145 | HTTPS FTP |
-Validation report
| Summary document |  emd_27145_validation.pdf.gz emd_27145_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27145_full_validation.pdf.gz emd_27145_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_27145_validation.xml.gz emd_27145_validation.xml.gz | 13 KB | Display | |
| Data in CIF |  emd_27145_validation.cif.gz emd_27145_validation.cif.gz | 15.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27145 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27145 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27145 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27145 | HTTPS FTP |
-Related structure data
| Related structure data |  8d2pMC  8d2kC  8d2lC  8d2nC  8d2oC  8d2qC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27145.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27145.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Accas9 ternary complex (Target bound) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
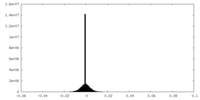
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cas9 ternary complex (Target bound)
| File | emd_27145_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cas9 ternary complex (Target bound) | ||||||||||||
| Projections & Slices |
| ||||||||||||
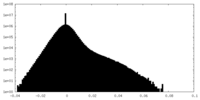
| Density Histograms |
-Half map: Halfmap2
| File | emd_27145_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
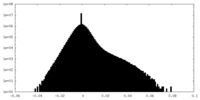
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM Structure of AceCas9 (Target bound)
| Entire | Name: CryoEM Structure of AceCas9 (Target bound) |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM Structure of AceCas9 (Target bound)
| Supramolecule | Name: CryoEM Structure of AceCas9 (Target bound) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: CRISPR-associated endonuclease, Csn1 family/RNA
| Supramolecule | Name: CRISPR-associated endonuclease, Csn1 family/RNA / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Acidothermus cellulolyticus 11B (bacteria) Acidothermus cellulolyticus 11B (bacteria) |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|
-Macromolecule #1: CRISPR-associated endonuclease, Csn1 family
| Macromolecule | Name: CRISPR-associated endonuclease, Csn1 family / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acidothermus cellulolyticus 11B (bacteria) / Strain: ATCC 43068 / DSM 8971 / 11B Acidothermus cellulolyticus 11B (bacteria) / Strain: ATCC 43068 / DSM 8971 / 11B |
| Molecular weight | Theoretical: 127.498094 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGGSEVGTVP VTWRLGVDVG ERSIGLAAVS YEEDKPKEIL AAVSWIHDGG VGDERSGASR LALRGMARRA RRLRRFRRAR LRDLDMLLS ELGWTPLPDK NVSPVDAWLA RKRLAEEYVV DETERRRLLG YAVSHMARHR GWRNPWTTIK DLKNLPQPSD S WERTRESL ...String: MGGSEVGTVP VTWRLGVDVG ERSIGLAAVS YEEDKPKEIL AAVSWIHDGG VGDERSGASR LALRGMARRA RRLRRFRRAR LRDLDMLLS ELGWTPLPDK NVSPVDAWLA RKRLAEEYVV DETERRRLLG YAVSHMARHR GWRNPWTTIK DLKNLPQPSD S WERTRESL EARYSVSLEP GTVGQWAGYL LQRAPGIRLN PTQQSAGRRA ELSNATAFET RLRQEDVLWE LRCIADVQGL PE DVVSNVI DAVFCQKRPS VPAERIGRDP LDPSQLRASR ACLEFQEYRI VAAVANLRIR DGSGSRPLSL EERNAVIEAL LAQ TERSLT WSDIALEILK LPNESDLTSV PEEDGPSSLA YSQFAPFDET SARIAEFIAK NRRKIPTFAQ WWQEQDRTSR SDLV AALAD NSIAGEEEQE LLVHLPDAEL EALEGLALPS GRVAYSRLTL SGLTRVMRDD GVDVHNARKT CFGVDDNWRP PLPAL HEAT GHPVVDRNLA ILRKFLSSAT MRWGPPQSIV VELARGASES RERQAEEEAA RRAHRKANDR IRAELRASGL SDPSPA DLV RARLLELYDC HCMYCGAPIS WENSELDHIV PRTDGGSNRH ENLAITCGAC NKEKGRRPFA SWAETSNRVQ LRDVIDR VQ KLKYSGNMYW TRDEFSRYKK SVVARLKRRT SDPEVIQSIE STGYAAVALR DRLLSYGEKN GVAQVAVFRG GVTAEARR W LDISIERLFS RVAIFAQSTS TKRLDRRHHA VDAVVLTTLT PGVAKTLADA RSRRVSAEFW RRPSDVNRHS TEEPQSPAY RQWKESCSGL GDLLISTAAR DSIAVAAPLR LRPTGALHEE TLRAFSEHTV GAAWKGAELR RIVEPEVYAA FLALTDPGGR FLKVSPSED VLPADENRHI VLSDRVLGPR DRVKLFPDDR GSIRVRGGAA YIASFHHARV FRWGSSHSPS FALLRVSLAD L AVAGLLRD GVDVFTAELP PWTPAWRYAS IALVKAVESG DAKQVGWLVP GDELDFGPEG VTTAAGDLSM FLKYFPERHW VV TGFEDDK RINLKPAFLS AEQAEVLRTE RSDRPDTLTE AGEILAQFFP RCWRATVAKV LCHPGLTVIR RTALGQPRWR RGH LPYSWR PWSADPWSGG TP UniProtKB: CRISPR-associated endonuclease, Csn1 family |
-Macromolecule #2: Single guide RNA (102-MER)
| Macromolecule | Name: Single guide RNA (102-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Acidothermus cellulolyticus 11B (bacteria) / Strain: ATCC 43068 / DSM 8971 / 11B Acidothermus cellulolyticus 11B (bacteria) / Strain: ATCC 43068 / DSM 8971 / 11B |
| Molecular weight | Theoretical: 34.390289 KDa |
| Sequence | String: (GTP)GUAGGAUGG CAAGAUCCUG GUAUGCUGGG GAGCCUGAAA AGGCUACCUA GCAAGACCCC UUCGUGGGGU CGCAUU CUU CACCCCCUCG CAGCAGCGAG GGGGUUC |
-Macromolecule #3: DNA target strand (5'-D(P*CP*CP*AP*GP*GP*AP*TP*CP*TP*TP*GP*CP*CP*...
| Macromolecule | Name: DNA target strand (5'-D(P*CP*CP*AP*GP*GP*AP*TP*CP*TP*TP*GP*CP*CP*AP*TP*CP*CP*TP*AP*CP*CP*TP*CP*T)-3') type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.216654 KDa |
| Sequence | String: (DC)(DC)(DA)(DG)(DG)(DA)(DT)(DC)(DT)(DT) (DG)(DC)(DC)(DA)(DT)(DC)(DC)(DT)(DA)(DC) (DC)(DT)(DC)(DT) |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 10 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 36 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.78 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 91324 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 4) |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)