[English] 日本語
 Yorodumi
Yorodumi- EMDB-26983: 1F8 mAb in complex with the computationally optimized broadly rea... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 1F8 mAb in complex with the computationally optimized broadly reactive H1 influenza hemagglutinin P1 | |||||||||
 Map data Map data | CryoEM map of COBRA P1 HA in complex with 1F8 mAb | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Hemagglutinin / Hemagglutinin /  influenza virus / influenza virus /  COBRA / COBRA /  monoclonal antibody / monoclonal antibody /  IMMUNE SYSTEM / IMMUNE SYSTEM /  Viral Protein-IMMUNE SYSTEM complex Viral Protein-IMMUNE SYSTEM complex | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /    Influenza A virus / Influenza A virus /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
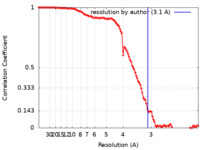
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.1 Å cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Dzimianski JV / DuBois RM / Ward A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Structural insights into the broad protection against H1 influenza viruses by a computationally optimized hemagglutinin vaccine. Authors: John V Dzimianski / Julianna Han / Giuseppe A Sautto / Sara M O'Rourke / Joseph M Cruz / Spencer R Pierce / Jeffrey W Ecker / Michael A Carlock / Kaito A Nagashima / Jarrod J Mousa / Ted M ...Authors: John V Dzimianski / Julianna Han / Giuseppe A Sautto / Sara M O'Rourke / Joseph M Cruz / Spencer R Pierce / Jeffrey W Ecker / Michael A Carlock / Kaito A Nagashima / Jarrod J Mousa / Ted M Ross / Andrew B Ward / Rebecca M DuBois /  Abstract: Influenza virus poses an ongoing human health threat with pandemic potential. Due to mutations in circulating strains, formulating effective vaccines remains a challenge. The use of computationally ...Influenza virus poses an ongoing human health threat with pandemic potential. Due to mutations in circulating strains, formulating effective vaccines remains a challenge. The use of computationally optimized broadly reactive antigen (COBRA) hemagglutinin (HA) proteins is a promising vaccine strategy to protect against a wide range of current and future influenza viruses. Though effective in preclinical studies, the mechanistic basis driving the broad reactivity of COBRA proteins remains to be elucidated. Here, we report the crystal structure of the COBRA HA termed P1 and identify antigenic and glycosylation properties that contribute to its immunogenicity. We further report the cryo-EM structure of the P1-elicited broadly neutralizing antibody 1F8 bound to COBRA P1, revealing 1F8 to recognize an atypical receptor binding site epitope via an unexpected mode of binding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26983.map.gz emd_26983.map.gz | 118.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26983-v30.xml emd-26983-v30.xml emd-26983.xml emd-26983.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26983_fsc.xml emd_26983_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_26983.png emd_26983.png | 74.9 KB | ||
| Masks |  emd_26983_msk_1.map emd_26983_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26983.cif.gz emd-26983.cif.gz | 6.7 KB | ||
| Others |  emd_26983_half_map_1.map.gz emd_26983_half_map_1.map.gz emd_26983_half_map_2.map.gz emd_26983_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26983 http://ftp.pdbj.org/pub/emdb/structures/EMD-26983 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26983 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26983 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26983.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26983.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of COBRA P1 HA in complex with 1F8 mAb | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26983_msk_1.map emd_26983_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
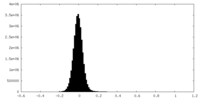
| Density Histograms |
-Half map: CryoEM half map B of COBRA P1 HA in complex with 1F8 mAb
| File | emd_26983_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM half map B of COBRA P1 HA in complex with 1F8 mAb | ||||||||||||
| Projections & Slices |
| ||||||||||||
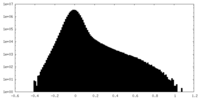
| Density Histograms |
-Half map: CryoEM half map A of COBRA P1 HA in complex with 1F8 mAb
| File | emd_26983_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM half map A of COBRA P1 HA in complex with 1F8 mAb | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : COBRA P1 HA in complex with 1F8 mAb
| Entire | Name: COBRA P1 HA in complex with 1F8 mAb |
|---|---|
| Components |
|
-Supramolecule #1: COBRA P1 HA in complex with 1F8 mAb
| Supramolecule | Name: COBRA P1 HA in complex with 1F8 mAb / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: COBRA P1 hemagglutinin
| Supramolecule | Name: COBRA P1 hemagglutinin / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: HA antigen designed from human H1N1 viruses from 1933-1957 and 2009-2011, and swine H1N1 viruses from 1931-1998. |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
-Supramolecule #3: 1F8 monoclonal antibody
| Supramolecule | Name: 1F8 monoclonal antibody / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 Details: Recombinantly expressed Fab fragment of an IgG antibody. |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
-Macromolecule #1: COBRA P1 HA
| Macromolecule | Name: COBRA P1 HA / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 63.264535 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DTICIGYHAN NSTDTVDTVL EKNVTVTHSV NLLEDSHNGK LCKLKGIAPL QLGKCNIAGW LLGNPECESL LSARSWSYIV ETPNSENGT CYPGDFIDYE ELREQLSSVS SFERFEIFPK ESSWPNHNTT KGVTAACSHA GKSSFYRNLL WLTKKGGSYP K LSKSYVNN ...String: DTICIGYHAN NSTDTVDTVL EKNVTVTHSV NLLEDSHNGK LCKLKGIAPL QLGKCNIAGW LLGNPECESL LSARSWSYIV ETPNSENGT CYPGDFIDYE ELREQLSSVS SFERFEIFPK ESSWPNHNTT KGVTAACSHA GKSSFYRNLL WLTKKGGSYP K LSKSYVNN KGKEVLVLWG VHHPSTSTDQ QSLYQNENAY VSVVSSNYNR RFTPEIAERP KVRGQAGRMN YYWTLLEPGD TI IFEATGN LIAPWYAFAL SRGSGSGIIT SNASMHECNT KCQTPQGAIN SSLPFQNIHP VTIGECPKYV RSTKLRMVTG LRN IPSIQS RGLFGAIAGF IEGGWTGMID GWYGYHHQNE QGSGYAADQK STQNAINGIT NKVNSVIEKM NTQFTAVGKE FNNL EKRME NLNKKVDDGF LDIWTYNAEL LVLLENERTL DFHDSNVKNL YEKVKSQLRN NAKEIGNGCF EFYHKCDNEC MESVK NGTY DYPKYSEESK LNREKIDGVK LESMGVYGSG YIPEAPRDGQ AYVRKDGEWV LLSTFLGLND IFEAQKIEWH EGHHHH HH |
-Macromolecule #2: 1F8 light chain
| Macromolecule | Name: 1F8 light chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 23.918271 KDa |
| Recombinant expression | Organism:   Cricetulus griseus (Chinese hamster) Cricetulus griseus (Chinese hamster) |
| Sequence | String: DIVLTQSPAS LAVSLGQRAT ISCKASQSVD FDGDTYMSWY QQKPGQPPKL LIYAASKLES GIPARFSGSG SGTDFTLNIH PVEEEDAAT YFCQQSNEDP WTFGGGTKLE IKRADAAPTV SIFPPSSEQL TSGGASVVCF LNNFYPKDIN VKWKIDGSER Q NGVLNSWT ...String: DIVLTQSPAS LAVSLGQRAT ISCKASQSVD FDGDTYMSWY QQKPGQPPKL LIYAASKLES GIPARFSGSG SGTDFTLNIH PVEEEDAAT YFCQQSNEDP WTFGGGTKLE IKRADAAPTV SIFPPSSEQL TSGGASVVCF LNNFYPKDIN VKWKIDGSER Q NGVLNSWT DQDSKDSTYS MSSTLTLTKD EYERHNSYTC EATHKTSTSP IVKSFNRNEC |
-Macromolecule #3: 1F8 heavy chain
| Macromolecule | Name: 1F8 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 23.900818 KDa |
| Recombinant expression | Organism:   Cricetulus griseus (Chinese hamster) Cricetulus griseus (Chinese hamster) |
| Sequence | String: QVQLQQSGAE LMKPGASVKI SCKATGYTFS SYWIEWVKQR PGHGLEWIGE ILPGSGRTNY DERFKGKATF TADTSSNTAY MQLSSLTSE DSAVYYCARP RIYGMDYWGQ GTSVTVSSAK TTPPSVYPLA PGSAAQTNSM VTLGCLVKGY FPEPVTVTWN S GSLSSGVH ...String: QVQLQQSGAE LMKPGASVKI SCKATGYTFS SYWIEWVKQR PGHGLEWIGE ILPGSGRTNY DERFKGKATF TADTSSNTAY MQLSSLTSE DSAVYYCARP RIYGMDYWGQ GTSVTVSSAK TTPPSVYPLA PGSAAQTNSM VTLGCLVKGY FPEPVTVTWN S GSLSSGVH TFPAVLQSDL YTLSSSVTVP SSPRPSETVT CNVAHPASST KVDKKIEPRG LVPR |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 10 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: TBS |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
| Details | 1:3:3 molar ratio of P1 HA, 1F8, and P1-05 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X