+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the sodium/iodide symporter (NIS) | |||||||||
 Map data Map data | Sharpened map after local refinement. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsodium:iodide symporter activity / iodide transmembrane transport / Thyroxine biosynthesis / salt transmembrane transporter activity / cellular response to Thyroid stimulating hormone / Organic anion transporters / monoatomic anion:sodium symporter activity / iodide transmembrane transporter activity / iodide transport / cellular response to gonadotropin stimulus ...sodium:iodide symporter activity / iodide transmembrane transport / Thyroxine biosynthesis / salt transmembrane transporter activity / cellular response to Thyroid stimulating hormone / Organic anion transporters / monoatomic anion:sodium symporter activity / iodide transmembrane transporter activity / iodide transport / cellular response to gonadotropin stimulus / symporter activity / thyroid hormone generation / sodium ion transport / cellular response to cAMP / cellular response to forskolin / transmembrane transport / protein homodimerization activity / extracellular exosome / nucleus / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Ravera S / Nicola JP / Salazar-De Simone G / Sigworth F / Karakas E / Amzel LM / Bianchet M / Carrasco N | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structural insights into the mechanism of the sodium/iodide symporter. Authors: Silvia Ravera / Juan Pablo Nicola / Glicella Salazar-De Simone / Fred J Sigworth / Erkan Karakas / L Mario Amzel / Mario A Bianchet / Nancy Carrasco /   Abstract: The sodium/iodide symporter (NIS) is the essential plasma membrane protein that mediates active iodide (I) transport into the thyroid gland, the first step in the biosynthesis of the thyroid hormones- ...The sodium/iodide symporter (NIS) is the essential plasma membrane protein that mediates active iodide (I) transport into the thyroid gland, the first step in the biosynthesis of the thyroid hormones-the master regulators of intermediary metabolism. NIS couples the inward translocation of I against its electrochemical gradient to the inward transport of Na down its electrochemical gradient. For nearly 50 years before its molecular identification, NIS was the molecule at the centre of the single most effective internal radiation cancer therapy: radioiodide (I) treatment for thyroid cancer. Mutations in NIS cause congenital hypothyroidism, which must be treated immediately after birth to prevent stunted growth and cognitive deficiency. Here we report three structures of rat NIS, determined by single-particle cryo-electron microscopy: one with no substrates bound; one with two Na and one I bound; and one with one Na and the oxyanion perrhenate bound. Structural analyses, functional characterization and computational studies show the substrate-binding sites and key residues for transport activity. Our results yield insights into how NIS selects, couples and translocates anions-thereby establishing a framework for understanding NIS function-and how it transports different substrates with different stoichiometries and releases substrates from its substrate-binding cavity into the cytosol. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26806.map.gz emd_26806.map.gz | 86 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26806-v30.xml emd-26806-v30.xml emd-26806.xml emd-26806.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
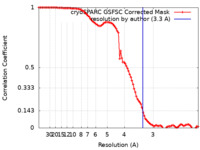
| FSC (resolution estimation) |  emd_26806_fsc.xml emd_26806_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_26806.png emd_26806.png | 74.3 KB | ||
| Masks |  emd_26806_msk_1.map emd_26806_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Others |  emd_26806_additional_1.map.gz emd_26806_additional_1.map.gz emd_26806_half_map_1.map.gz emd_26806_half_map_1.map.gz emd_26806_half_map_2.map.gz emd_26806_half_map_2.map.gz | 45.6 MB 84.7 MB 84.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26806 http://ftp.pdbj.org/pub/emdb/structures/EMD-26806 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26806 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26806 | HTTPS FTP |
-Validation report
| Summary document |  emd_26806_validation.pdf.gz emd_26806_validation.pdf.gz | 717 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26806_full_validation.pdf.gz emd_26806_full_validation.pdf.gz | 716.5 KB | Display | |
| Data in XML |  emd_26806_validation.xml.gz emd_26806_validation.xml.gz | 17.9 KB | Display | |
| Data in CIF |  emd_26806_validation.cif.gz emd_26806_validation.cif.gz | 22.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26806 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26806 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26806 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26806 | HTTPS FTP |
-Related structure data
| Related structure data |  7uuyMC  7uuzC  7uv0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26806.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26806.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map after local refinement. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.068 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26806_msk_1.map emd_26806_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map after local refinement
| File | emd_26806_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map after local refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_26806_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26806_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Sodium/iodide symporter
| Entire | Name: Sodium/iodide symporter |
|---|---|
| Components |
|
-Supramolecule #1: Sodium/iodide symporter
| Supramolecule | Name: Sodium/iodide symporter / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 75 KDa |
-Macromolecule #1: Sodium/iodide cotransporter
| Macromolecule | Name: Sodium/iodide cotransporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 73.914414 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MYPYDVPDYA ENLYFQSEGA EAGARATFGA WDYGVFATML LVSTGIGLWV GLARGGQRSA DDFFTGGRQL AAVPVGLSLA ASFMSAVQV LGVPAEAARY GLKFLWMCAG QLLNSLLTAF LFLPIFYRLG LTSTYQYLEL RFSRAVRLCG TLQYLVATML Y TGIVIYAP ...String: MYPYDVPDYA ENLYFQSEGA EAGARATFGA WDYGVFATML LVSTGIGLWV GLARGGQRSA DDFFTGGRQL AAVPVGLSLA ASFMSAVQV LGVPAEAARY GLKFLWMCAG QLLNSLLTAF LFLPIFYRLG LTSTYQYLEL RFSRAVRLCG TLQYLVATML Y TGIVIYAP ALILNQVTGL DIWASLLSTG IICTLYTTVG GMKAVVWTDV FQVVVMLVGF WVILARGVIL LGGPRNVLSL AQ QHSRINL MDFDPDPRSR YTFWTFIVGG TLVWLSMYGV NQAQVQRYVA CHTEGKAKLA LLVNQLGLFL IVASAACCGI VMF VYYKDC DPLLTGRISA PDQYMPLLVL DIFEDLPGVP GLFLACAYSG TLSTASTSIN AMAAVTVEDL IKPRMPGLAP RKLV FISKG LSFIYGSACL TVAALSSLLG GGVLQGSFTV MGVISGPLLG AFTLGMLLPA CNTPGVLSGL AAGLAVSLWV AVGAT LYPP GEQTMGVLPT SAAGCTQDSV LLGPPGATQA SNGIPSSGMD TGRPALADTF YAISYLYYGA LGTLTTMLCG ALISYL TGP TKRSSLGPGL LWWDLARQTA SVAPKEDTAT LEESLVKGPE DIPAVTKKPP GLKPGAETHP LYLGHDVETN LSGGGGA LE VLFQGPHHHH HHHHMDEKTT GWRGGHVVEG LAGELEQLRA RLEHHPQGQR EP |
-Macromolecule #2: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE
| Macromolecule | Name: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: 3PH |
|---|---|
| Molecular weight | Theoretical: 704.998 Da |
| Chemical component information |  ChemComp-3PH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Sugar embedding | Material: vitrified ice | |||||||||||||||
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 2 / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7uuy: |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X