[English] 日本語
 Yorodumi
Yorodumi- EMDB-26712: cryo-EM structure of ribonucleotide reductase from Synechococcus ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of ribonucleotide reductase from Synechococcus phage S-CBP4 bound with TTP | |||||||||
 Map data Map data | Sharpened map from the reconstruction with C2 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribonucleotide reductase / Synechoccus phage / TTP / OXIDOREDUCTASE | |||||||||
| Function / homology | : / Ribonucleotide reductase large subunit, C-terminal / Ribonucleotide reductase, barrel domain / cobalamin binding / ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor / Ribonucleotide reductase domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |  Synechococcus phage S-CBP4 (virus) Synechococcus phage S-CBP4 (virus) | |||||||||
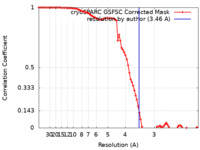
| Method | single particle reconstruction / cryo EM / Resolution: 3.46 Å | |||||||||
 Authors Authors | Xu D / Burnim AA / Ando N | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Comprehensive phylogenetic analysis of the ribonucleotide reductase family reveals an ancestral clade. Authors: Audrey A Burnim / Matthew A Spence / Da Xu / Colin J Jackson / Nozomi Ando /   Abstract: Ribonucleotide reductases (RNRs) are used by all free-living organisms and many viruses to catalyze an essential step in the de novo biosynthesis of DNA precursors. RNRs are remarkably diverse by ...Ribonucleotide reductases (RNRs) are used by all free-living organisms and many viruses to catalyze an essential step in the de novo biosynthesis of DNA precursors. RNRs are remarkably diverse by primary sequence and cofactor requirement, while sharing a conserved fold and radical-based mechanism for nucleotide reduction. Here, we structurally aligned the diverse RNR family by the conserved catalytic barrel to reconstruct the first large-scale phylogeny consisting of 6779 sequences that unites all extant classes of the RNR family and performed evo-velocity analysis to independently validate our evolutionary model. With a robust phylogeny in-hand, we uncovered a novel, phylogenetically distinct clade that is placed as ancestral to the classes I and II RNRs, which we have termed clade Ø. We employed small-angle X-ray scattering (SAXS), cryogenic-electron microscopy (cryo-EM), and AlphaFold2 to investigate a member of this clade from phage S-CBP4 and report the most minimal RNR architecture to-date. Based on our analyses, we propose an evolutionary model of diversification in the RNR family and delineate how our phylogeny can be used as a roadmap for targeted future study. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26712.map.gz emd_26712.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26712-v30.xml emd-26712-v30.xml emd-26712.xml emd-26712.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26712_fsc.xml emd_26712_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_26712.png emd_26712.png | 70.9 KB | ||
| Filedesc metadata |  emd-26712.cif.gz emd-26712.cif.gz | 7.3 KB | ||
| Others |  emd_26712_half_map_1.map.gz emd_26712_half_map_1.map.gz emd_26712_half_map_2.map.gz emd_26712_half_map_2.map.gz | 59.2 MB 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26712 http://ftp.pdbj.org/pub/emdb/structures/EMD-26712 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26712 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26712 | HTTPS FTP |
-Validation report
| Summary document |  emd_26712_validation.pdf.gz emd_26712_validation.pdf.gz | 757 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26712_full_validation.pdf.gz emd_26712_full_validation.pdf.gz | 756.6 KB | Display | |
| Data in XML |  emd_26712_validation.xml.gz emd_26712_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_26712_validation.cif.gz emd_26712_validation.cif.gz | 21.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26712 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26712 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26712 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26712 | HTTPS FTP |
-Related structure data
| Related structure data |  7urgMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26712.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26712.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map from the reconstruction with C2 symmetry | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A from the reconstruction with C2 symmetry
| File | emd_26712_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A from the reconstruction with C2 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B from the reconstruction with C2 symmetry
| File | emd_26712_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B from the reconstruction with C2 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ribonucleotide reductase from Synechoccus phage S-CBP4 bound with TTP
| Entire | Name: Ribonucleotide reductase from Synechoccus phage S-CBP4 bound with TTP |
|---|---|
| Components |
|
-Supramolecule #1: Ribonucleotide reductase from Synechoccus phage S-CBP4 bound with TTP
| Supramolecule | Name: Ribonucleotide reductase from Synechoccus phage S-CBP4 bound with TTP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Synechococcus phage S-CBP4 (virus) Synechococcus phage S-CBP4 (virus) |
-Macromolecule #1: Ribonucleotide reductase
| Macromolecule | Name: Ribonucleotide reductase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Synechococcus phage S-CBP4 (virus) Synechococcus phage S-CBP4 (virus) |
| Molecular weight | Theoretical: 51.421672 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKPPKELIA RTGRVQSWID DPTSRLPVSC TVFVVEDTME GENGIEASWR FVSHALRYGA GVAVHLSKLR PKGAENGKGL VASGPVSFA KIYSTLNEIL RRGGVYKNGA VVCHLDLSHP DVLEFITASR SELPWVKRCV NINDHWWKEA TPTVKNALLE G IKRGDIWL ...String: MSKPPKELIA RTGRVQSWID DPTSRLPVSC TVFVVEDTME GENGIEASWR FVSHALRYGA GVAVHLSKLR PKGAENGKGL VASGPVSFA KIYSTLNEIL RRGGVYKNGA VVCHLDLSHP DVLEFITASR SELPWVKRCV NINDHWWKEA TPTVKNALLE G IKRGDIWL NKTKVDRNGN RIRGNVCLEV YLPSRGTCLL QHVNLGGCEL DEIRGAFAQG MSELCELHGK TNVGESGEYL PS ETDRQVG LGMLGLANLL RTQGVTYNDF GRALEALNSG RPYPSTPGYV IAQELKAGIQ AAAEIAKANK MERAFAIAPT ASC SYRYTD LDGYTTCPEI APPIARQVDR DSGTFGVQSF DYGPVEIASE VGWESYKRVV DGIIRLLDST GLLHGYSFNS WSDV VTYDE QFIEDWLASP QTSLYYSLQV MGDVQDKSDA YAALDDGDVT AYLESLLNDP VGASPPLAPD CNCGE UniProtKB: Ribonucleotide reductase domain-containing protein |
-Macromolecule #2: THYMIDINE-5'-TRIPHOSPHATE
| Macromolecule | Name: THYMIDINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: TTP |
|---|---|
| Molecular weight | Theoretical: 482.168 Da |
| Chemical component information |  ChemComp-TTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
Details: 50 mM HEPES, 150 mM NaCl, 1% v/v glycerol, 7.55 mM MgCl2 | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 0.039 kPa Details: glow discharged on a PELCO easiGlow system for 45 s with 15 mA current | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 4 seconds before plunging. | |||||||||||||||
| Details | 4 uM ribonucleotide reductase from Synechococcus phage S-CBP4 with 200 uM TTP, 200 uM GDP |
- Electron microscopy
Electron microscopy
| Microscope | TFS TALOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Software | Name: SerialEM (ver. 3.8) |
| Details | Data was collected on a Thermo Fisher Talos Arcica Cryo-TEM with a Gatan K3 camera and BioQuantum energy filter. |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 856 / Average exposure time: 2.164 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 79000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Software | Name: PHENIX (ver. 1.20.1) Details: PHENIX was used to dock the alpha fold model into the EM map. |
|---|---|
| Details | The sequence for the ribonucleotide reductase from Synechococcus phage S-CBP4 was retrieved from UniProt with accession number M1PRZ0. The sequence was used as input for AlphaFold2 prediction with the five default model parameters and a template date cutoff of 2020-05-14. As the five models were largely identical in the core region and differing only in the location of the C-terminal tail, the structure predicted with the first model parameter was used in the subsequent process. The predicted structure was first processed and docked into the unsharpened map in phenix. The 25 N-terminal residues and 45 C-terminal residues were then manually removed due to lack of cryo-EM density, and residues 26-426 were retained in the model. We observed unmodeled density at the specificity site, and based on solution composition, we modeled a TTP molecule. The TTP molecule with magnesium ion from the crystal structure of Bacillus subtilis RNR (pdb: 6mt9) was extracted and rigid body fit into the unmodeled density in Coot. The combined model was refined with the unsharpened and sharpened maps using phenix.real_space_refine, with a constraint applied on the magnesium ion coordinated by the triphosphate in TTP according to the original configuration. Residue and loop conformations in the resulting structure were manually adjusted in Coot to maximize fit to map and input for an additional round of real-space refinement in phenix with an additional restraint for the disulfide bond between C30 and C196. Due to poor density of the magnesium ion, it was removed when deposited into PDB. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 74.37 |
| Output model |  PDB-7urg: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)