[English] 日本語
 Yorodumi
Yorodumi- EMDB-26679: Subtomogram averaged map of hACE2 dimers on the surface of extrac... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
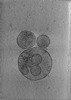
| Title | Subtomogram averaged map of hACE2 dimers on the surface of extracellular vesicles | ||||||||||||
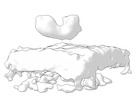
 Map data Map data | Final subtomogram averaged map of hACE2 on the surface of extracellular vesicles. | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
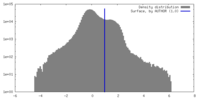
| Method | subtomogram averaging / cryo EM / Resolution: 19.0 Å | ||||||||||||
 Authors Authors | Marcink TC / Kicmal T / Armbruster E / Zhang Z / Zipursky G / Idris M / Khao J / McGill G / Gallagher T / Porotto M ...Marcink TC / Kicmal T / Armbruster E / Zhang Z / Zipursky G / Idris M / Khao J / McGill G / Gallagher T / Porotto M / des Georges A / Moscona A | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Intermediates in SARS-CoV-2 spike-mediated cell entry. Authors: Tara C Marcink / Thomas Kicmal / Emily Armbruster / Zhening Zhang / Gillian Zipursky / Kate L Golub / Mohab Idris / Jonathan Khao / Jennifer Drew-Bear / Gael McGill / Tom Gallagher / Matteo ...Authors: Tara C Marcink / Thomas Kicmal / Emily Armbruster / Zhening Zhang / Gillian Zipursky / Kate L Golub / Mohab Idris / Jonathan Khao / Jennifer Drew-Bear / Gael McGill / Tom Gallagher / Matteo Porotto / Amédée des Georges / Anne Moscona /   Abstract: SARS-CoV-2 cell entry is completed after viral spike (S) protein-mediated membrane fusion between viral and host cell membranes. Stable prefusion and postfusion S structures have been resolved by ...SARS-CoV-2 cell entry is completed after viral spike (S) protein-mediated membrane fusion between viral and host cell membranes. Stable prefusion and postfusion S structures have been resolved by cryo-electron microscopy and cryo-electron tomography, but the refolding intermediates on the fusion pathway are transient and have not been examined. We used an antiviral lipopeptide entry inhibitor to arrest S protein refolding and thereby capture intermediates as S proteins interact with hACE2 and fusion-activating proteases on cell-derived target membranes. Cryo-electron tomography imaged both extended and partially folded intermediate states of S2, as well as a novel late-stage conformation on the pathway to membrane fusion. The intermediates now identified in this dynamic S protein-directed fusion provide mechanistic insights that may guide the design of CoV entry inhibitors. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26679.map.gz emd_26679.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26679-v30.xml emd-26679-v30.xml emd-26679.xml emd-26679.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26679.png emd_26679.png | 39.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26679 http://ftp.pdbj.org/pub/emdb/structures/EMD-26679 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26679 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26679 | HTTPS FTP |
-Validation report
| Summary document |  emd_26679_validation.pdf.gz emd_26679_validation.pdf.gz | 380.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26679_full_validation.pdf.gz emd_26679_full_validation.pdf.gz | 380 KB | Display | |
| Data in XML |  emd_26679_validation.xml.gz emd_26679_validation.xml.gz | 5.5 KB | Display | |
| Data in CIF |  emd_26679_validation.cif.gz emd_26679_validation.cif.gz | 6.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26679 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26679 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26679 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26679 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26679.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26679.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final subtomogram averaged map of hACE2 on the surface of extracellular vesicles. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.24 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Dimeric complex consisting of full-length membrane-bound human AC...
| Entire | Name: Dimeric complex consisting of full-length membrane-bound human ACE2 with a LgBiT tag. |
|---|---|
| Components |
|
-Supramolecule #1: Dimeric complex consisting of full-length membrane-bound human AC...
| Supramolecule | Name: Dimeric complex consisting of full-length membrane-bound human ACE2 with a LgBiT tag. type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293T / Recombinant plasmid: pcDNA3.1 Homo sapiens (human) / Recombinant cell: HEK293T / Recombinant plasmid: pcDNA3.1 |
-Macromolecule #1: hACE2-LgBiT
| Macromolecule | Name: hACE2-LgBiT / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSSSSWLLLS LVAVTAAQST IEEQAKTFLD KFNHEAEDLF YQSSLASWNY NTNITEENVQ NMNNAGDKWS AFLKEQSTLA QMYPLQEIQN LTVKLQLQAL QQNGSSVLSE DKSKRLNTIL NTMSTIYSTG KVCNPDNPQE CLLLEPGLNE IMANSLDYNE RLWAWESWRS ...String: MSSSSWLLLS LVAVTAAQST IEEQAKTFLD KFNHEAEDLF YQSSLASWNY NTNITEENVQ NMNNAGDKWS AFLKEQSTLA QMYPLQEIQN LTVKLQLQAL QQNGSSVLSE DKSKRLNTIL NTMSTIYSTG KVCNPDNPQE CLLLEPGLNE IMANSLDYNE RLWAWESWRS EVGKQLRPLY EEYVVLKNEM ARANHYEDYG DYWRGDYEVN GVDGYDYSRG QLIEDVEHTF EEIKPLYEHL HAYVRAKLMN AYPSYISPIG CLPAHLLGDM WGRFWTNLYS LTVPFGQKPN IDVTDAMVDQ AWDAQRIFKE AEKFFVSVGL PNMTQGFWEN SMLTDPGNVQ KAVCHPTAWD LGKGDFRILM CTKVTMDDFL TAHHEMGHIQ YDMAYAAQPF LLRNGANEGF HEAVGEIMSL SAATPKHLKS IGLLSPDFQE DNETEINFLL KQALTIVGTL PFTYMLEKWR WMVFKGEIPK DQWMKKWWEM KREIVGVVEP VPHDETYCDP ASLFHVSNDY SFIRYYTRTL YQFQFQEALC QAAKHEGPLH KCDISNSTEA GQKLFNMLRL GKSEPWTLAL ENVVGAKNMN VRPLLNYFEP LFTWLKDQNK NSFVGWSTDW SPYADQSIKV RISLKSALGD KAYEWNDNEM YLFRSSVAYA MRQYFLKVKN QMILFGEEDV RVANLKPRIS FNFFVTAPKN VSDIIPRTEV EKAIRMSRSR INDAFRLNDN SLEFLGIQPT LGPPNQPPVS IWLIVFGVVM GVIVVGIVIL IFTGIRDRKK KNKARSGENP YASIDISKGE NNPGFQNTDD VQTSF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3.0 nm |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum SE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 2.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 8.0 µm / Nominal defocus min: 4.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 19.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: Dynamo (ver. 1.1.514) / Number subtomograms used: 315 |
|---|---|
| Extraction | Number tomograms: 1 / Number images used: 1 |
| CTF correction | Software - Name: Warp (ver. 1.0.9) |
| Final angle assignment | Type: NOT APPLICABLE |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)