[English] 日本語
 Yorodumi
Yorodumi- EMDB-26646: SARS-CoV-2 replication-transcription complex bound to CTP, in a p... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state | |||||||||
 Map data Map data | SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA-directed 5'-3' RNA polymerase activity / positive stranded viral RNA replication / REPLICATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / mRNA guanylyltransferase activity / 5'-3' RNA helicase activity / Lyases; Phosphorus-oxygen lyases / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / Maturation of replicase proteins / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / ISG15-specific peptidase activity / TRAF3-dependent IRF activation pathway ...protein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / mRNA guanylyltransferase activity / 5'-3' RNA helicase activity / Lyases; Phosphorus-oxygen lyases / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / Maturation of replicase proteins / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / ISG15-specific peptidase activity / TRAF3-dependent IRF activation pathway / Transcription of SARS-CoV-2 sgRNAs / Translation of Replicase and Assembly of the Replication Transcription Complex / snRNP Assembly / Replication of the SARS-CoV-2 genome / double membrane vesicle viral factory outer membrane / Hydrolases; Acting on ester bonds; Exoribonucleases producing 5'-phosphomonoesters / host cell endoplasmic reticulum-Golgi intermediate compartment / SARS coronavirus main proteinase / 3'-5'-RNA exonuclease activity / 5'-3' DNA helicase activity / host cell endosome / symbiont-mediated degradation of host mRNA / mRNA guanylyltransferase / symbiont-mediated suppression of host ISG15-protein conjugation / G-quadruplex RNA binding / symbiont-mediated suppression of host toll-like receptor signaling pathway / omega peptidase activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / SARS-CoV-2 modulates host translation machinery / mRNA (guanine-N7)-methyltransferase / host cell Golgi apparatus / methyltransferase cap1 / symbiont-mediated perturbation of host ubiquitin-like protein modification / symbiont-mediated suppression of host NF-kappaB cascade / DNA helicase / methyltransferase cap1 activity / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / forked DNA-dependent helicase activity / single-stranded 3'-5' DNA helicase activity / four-way junction helicase activity / double-stranded DNA helicase activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / single-stranded RNA binding / chromatin extrusion motor activity / ATP-dependent H2AZ histone chaperone activity / cohesin loader activity / ATP-dependent H3-H4 histone complex chaperone activity / DNA clamp loader activity / host cell perinuclear region of cytoplasm / viral protein processing / lyase activity / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / : / copper ion binding / viral translational frameshifting / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / symbiont-mediated suppression of host gene expression / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / lipid binding / host cell nucleus / SARS-CoV-2 activates/modulates innate and adaptive immune responses / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.67 Å | |||||||||
 Authors Authors | Malone BF / Perry JK / Appleby TC / Feng JY / Campbell EA / Darst SA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structural basis for substrate selection by the SARS-CoV-2 replicase. Authors: Brandon F Malone / Jason K Perry / Paul Dominic B Olinares / Hery W Lee / James Chen / Todd C Appleby / Joy Y Feng / John P Bilello / Honkit Ng / Johanna Sotiris / Mark Ebrahim / Eugene Y D ...Authors: Brandon F Malone / Jason K Perry / Paul Dominic B Olinares / Hery W Lee / James Chen / Todd C Appleby / Joy Y Feng / John P Bilello / Honkit Ng / Johanna Sotiris / Mark Ebrahim / Eugene Y D Chua / Joshua H Mendez / Ed T Eng / Robert Landick / Matthias Götte / Brian T Chait / Elizabeth A Campbell / Seth A Darst /   Abstract: The SARS-CoV-2 RNA-dependent RNA polymerase coordinates viral RNA synthesis as part of an assembly known as the replication-transcription complex (RTC). Accordingly, the RTC is a target for ...The SARS-CoV-2 RNA-dependent RNA polymerase coordinates viral RNA synthesis as part of an assembly known as the replication-transcription complex (RTC). Accordingly, the RTC is a target for clinically approved antiviral nucleoside analogues, including remdesivir. Faithful synthesis of viral RNAs by the RTC requires recognition of the correct nucleotide triphosphate (NTP) for incorporation into the nascent RNA. To be effective inhibitors, antiviral nucleoside analogues must compete with the natural NTPs for incorporation. How the SARS-CoV-2 RTC discriminates between the natural NTPs, and how antiviral nucleoside analogues compete, has not been discerned in detail. Here, we use cryogenic-electron microscopy to visualize the RTC bound to each of the natural NTPs in states poised for incorporation. Furthermore, we investigate the RTC with the active metabolite of remdesivir, remdesivir triphosphate (RDV-TP), highlighting the structural basis for the selective incorporation of RDV-TP over its natural counterpart adenosine triphosphate. Our results explain the suite of interactions required for NTP recognition, informing the rational design of antivirals. Our analysis also yields insights into nucleotide recognition by the nsp12 NiRAN (nidovirus RdRp-associated nucleotidyltransferase), an enigmatic catalytic domain essential for viral propagation. The NiRAN selectively binds guanosine triphosphate, strengthening proposals for the role of this domain in the formation of the 5' RNA cap. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26646.map.gz emd_26646.map.gz | 108.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26646-v30.xml emd-26646-v30.xml emd-26646.xml emd-26646.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26646_fsc.xml emd_26646_fsc.xml | 13.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_26646.png emd_26646.png | 40.7 KB | ||
| Filedesc metadata |  emd-26646.cif.gz emd-26646.cif.gz | 6.9 KB | ||
| Others |  emd_26646_additional_1.map.gz emd_26646_additional_1.map.gz emd_26646_half_map_1.map.gz emd_26646_half_map_1.map.gz emd_26646_half_map_2.map.gz emd_26646_half_map_2.map.gz | 8.2 MB 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26646 http://ftp.pdbj.org/pub/emdb/structures/EMD-26646 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26646 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26646 | HTTPS FTP |
-Validation report
| Summary document |  emd_26646_validation.pdf.gz emd_26646_validation.pdf.gz | 780.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26646_full_validation.pdf.gz emd_26646_full_validation.pdf.gz | 779.7 KB | Display | |
| Data in XML |  emd_26646_validation.xml.gz emd_26646_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_26646_validation.cif.gz emd_26646_validation.cif.gz | 28.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26646 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26646 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26646 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26646 | HTTPS FTP |
-Related structure data
| Related structure data |  7uoeMC  7uo4C  7uo7C  7uo9C  7uobC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26646.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26646.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||
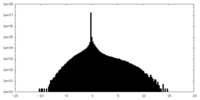
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Local resolution filtered map
| File | emd_26646_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
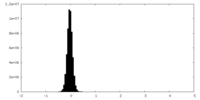
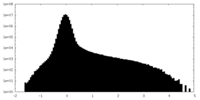
| Density Histograms |
-Half map: SARS-CoV-2 replication-transcription complex bound to CTP, in a...
| File | emd_26646_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state | ||||||||||||
| Projections & Slices |
| ||||||||||||
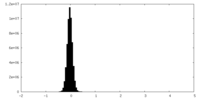
| Density Histograms |
-Half map: SARS-CoV-2 replication-transcription complex bound to CTP, in a...
| File | emd_26646_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : SARS-CoV-2 replication-transcription complex + CTP
+Supramolecule #1: SARS-CoV-2 replication-transcription complex + CTP
+Macromolecule #1: RNA-directed RNA polymerase
+Macromolecule #2: Non-structural protein 8
+Macromolecule #3: Non-structural protein 7
+Macromolecule #4: Product RNA (35-MER)
+Macromolecule #5: Template RNA (55-MER)
+Macromolecule #6: ZINC ION
+Macromolecule #7: MAGNESIUM ION
+Macromolecule #8: CYTIDINE-5'-TRIPHOSPHATE
+Macromolecule #9: 3'-DEOXYURIDINE-5'-MONOPHOSPHATE
+Macromolecule #10: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 56.76 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)