[English] 日本語
 Yorodumi
Yorodumi- EMDB-26566: Bacteriophage Lambda Red-Beta N-terminal domain helical assembly ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacteriophage Lambda Red-Beta N-terminal domain helical assembly in complex with dsDNA | |||||||||
 Map data Map data | Symmetrised, sharpened map of the RedBeta177 helical assembly bound to dsDNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Annealase / Synaptase / SSAP / Single-strand annealing protein / DNA annealing intermediate / Recombinase / Two-component recombinase / Viral / DNA-binding / RECOMBINATION-DNA complex | |||||||||
| Function / homology | Bacteriophage lambda, Recombination protein bet / RecT family / RecT family / DNA recombination / DNA binding / Recombination protein bet Function and homology information Function and homology information | |||||||||
| Biological species |  Escherichia virus Lambda / Escherichia virus Lambda /  | |||||||||
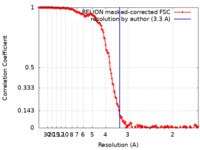
| Method | helical reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Newing TP / Tolun G | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Redβ annealase structure reveals details of oligomerization and λ Red-mediated homologous DNA recombination. Authors: Timothy P Newing / Jodi L Brewster / Lucy J Fitschen / James C Bouwer / Nikolas P Johnston / Haibo Yu / Gökhan Tolun /  Abstract: The Redβ protein of the bacteriophage λ red recombination system is a model annealase which catalyzes single-strand annealing homologous DNA recombination. Here we present the structure of a ...The Redβ protein of the bacteriophage λ red recombination system is a model annealase which catalyzes single-strand annealing homologous DNA recombination. Here we present the structure of a helical oligomeric annealing intermediate of Redβ, consisting of N-terminal residues 1-177 bound to two complementary 27mer oligonucleotides, determined via cryogenic electron microscopy (cryo-EM) to a final resolution of 3.3 Å. The structure reveals a continuous binding groove which positions and stabilizes complementary DNA strands in a planar orientation to facilitate base pairing via a network of hydrogen bonding. Definition of the inter-subunit interface provides a structural basis for the propensity of Redβ to oligomerize into functionally significant long helical filaments, a trait shared by most annealases. Our cryo-EM structure and molecular dynamics simulations suggest that residues 133-138 form a flexible loop which modulates access to the binding groove. More than half a century after its discovery, this combination of structural and computational observations has allowed us to propose molecular mechanisms for the actions of the model annealase Redβ, a defining member of the Redβ/RecT protein family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26566.map.gz emd_26566.map.gz | 106.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26566-v30.xml emd-26566-v30.xml emd-26566.xml emd-26566.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26566_fsc.xml emd_26566_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_26566.png emd_26566.png | 175.3 KB | ||
| Masks |  emd_26566_msk_1.map emd_26566_msk_1.map | 343 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26566.cif.gz emd-26566.cif.gz | 6.8 KB | ||
| Others |  emd_26566_half_map_1.map.gz emd_26566_half_map_1.map.gz emd_26566_half_map_2.map.gz emd_26566_half_map_2.map.gz | 318.4 MB 318.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26566 http://ftp.pdbj.org/pub/emdb/structures/EMD-26566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26566 | HTTPS FTP |
-Validation report
| Summary document |  emd_26566_validation.pdf.gz emd_26566_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26566_full_validation.pdf.gz emd_26566_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_26566_validation.xml.gz emd_26566_validation.xml.gz | 23.9 KB | Display | |
| Data in CIF |  emd_26566_validation.cif.gz emd_26566_validation.cif.gz | 31.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26566 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26566 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26566 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26566 | HTTPS FTP |
-Related structure data
| Related structure data |  7ujlMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26566.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26566.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Symmetrised, sharpened map of the RedBeta177 helical assembly bound to dsDNA | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26566_msk_1.map emd_26566_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half map B of the RedBeta177 helical...
| File | emd_26566_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map B of the RedBeta177 helical assembly bound to dsDNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half map A of the RedBeta177 helical...
| File | emd_26566_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map A of the RedBeta177 helical assembly bound to dsDNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RedBeta177 oligomeric helical assembly bound to two complementary...
| Entire | Name: RedBeta177 oligomeric helical assembly bound to two complementary 27mer ssDNA oligonucleotides |
|---|---|
| Components |
|
-Supramolecule #1: RedBeta177 oligomeric helical assembly bound to two complementary...
| Supramolecule | Name: RedBeta177 oligomeric helical assembly bound to two complementary 27mer ssDNA oligonucleotides type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Helical complex assembled through sequential addition of complementary oligonucleotides in a controlled environment |
|---|---|
| Molecular weight | Theoretical: 102.26 kDa/nm |
-Supramolecule #2: Red-beta annealase N-terminal domain
| Supramolecule | Name: Red-beta annealase N-terminal domain / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Escherichia virus Lambda Escherichia virus Lambda |
-Supramolecule #3: Template ssDNA
| Supramolecule | Name: Template ssDNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 / Details: 27mer ssDNA oligonucleotide |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #4: Complementary ssDNA
| Supramolecule | Name: Complementary ssDNA / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 / Details: 27mer ssDNA oligonucleotide |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Recombination protein bet
| Macromolecule | Name: Recombination protein bet / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia virus Lambda Escherichia virus Lambda |
| Molecular weight | Theoretical: 21.275008 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTALATLAG KLAERVGMDS VDPQELITTL RQTAFKGDAS DAQFIALLIV ANQYGLNPWT KEIYAFPDKQ NGIVPVVGVD GWSRIINEN QQFDGMDFEQ DNESCTCRIY RKDRNHPICV TEWMDECRRE PFKTREGREI TGPWQSHPKR MLRHKAMIQC A RLAFGFAG IYDKDEAERS SHHHHHH UniProtKB: Recombination protein bet |
-Macromolecule #2: Template DNA
| Macromolecule | Name: Template DNA / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.244295 KDa |
| Sequence | String: (DT)(DG)(DC)(DA)(DG)(DC)(DA)(DG)(DC)(DT) (DT)(DT)(DA)(DC)(DC)(DA)(DT)(DC)(DT)(DG) (DC)(DC)(DG)(DC)(DT)(DG)(DG) |
-Macromolecule #3: Complementary DNA
| Macromolecule | Name: Complementary DNA / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.351386 KDa |
| Sequence | String: (DC)(DC)(DA)(DG)(DC)(DG)(DG)(DC)(DA)(DG) (DA)(DT)(DG)(DG)(DT)(DA)(DA)(DA)(DG)(DC) (DT)(DG)(DC)(DT)(DG)(DC)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 2.6 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: Sample loading volume ranged between 2 and 3 microlitres. Samples were blotted for 5 seconds prior to vitrification.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Details: Installed but not used |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 4710 / Average exposure time: 9.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 59500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-7ujl: |
 Movie
Movie Controller
Controller



 Z
Z Y
Y X
X