+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoET of E. coli prepared with the Waffle Method | |||||||||||||||
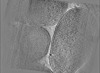
 Map data Map data | E. coli waffle lamella tomogram from Appion-Protomo alignment. Pixelsize is actually 8.30616; ignore the filename. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | electron tomography / cryo EM | |||||||||||||||
 Authors Authors | Kelley K / Raczkowski AM / Klykov O / Jaroenlak P / Bobe D / Kopylov M / Eng ET / Bhabha G / Potter CS / Carragher B / Noble AJ | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Waffle Method: A general and flexible approach for improving throughput in FIB-milling. Authors: Kotaro Kelley / Ashleigh M Raczkowski / Oleg Klykov / Pattana Jaroenlak / Daija Bobe / Mykhailo Kopylov / Edward T Eng / Gira Bhabha / Clinton S Potter / Bridget Carragher / Alex J Noble /  Abstract: Cryo-FIB/SEM combined with cryo-ET has emerged from within the field of cryo-EM as the method for obtaining the highest resolution structural information of complex biological samples in-situ in ...Cryo-FIB/SEM combined with cryo-ET has emerged from within the field of cryo-EM as the method for obtaining the highest resolution structural information of complex biological samples in-situ in native and non-native environments. However, challenges remain in conventional cryo-FIB/SEM workflows, including milling thick specimens with vitrification issues, specimens with preferred orientation, low-throughput when milling small and/or low concentration specimens, and specimens that distribute poorly across grid squares. Here we present a general approach called the 'Waffle Method' which leverages high-pressure freezing to address these challenges. We illustrate the mitigation of these challenges by applying the Waffle Method and cryo-ET to reveal the macrostructure of the polar tube in microsporidian spores in multiple complementary orientations, which was previously not possible due to preferred orientation. We demonstrate the broadness of the Waffle Method by applying it to three additional cellular samples and a single particle sample using a variety of cryo-FIB-milling hardware, with manual and automated approaches. We also present a unique and critical stress-relief gap designed specifically for waffled lamellae. We propose the Waffle Method as a way to achieve many advantages of cryo-liftout on the specimen grid while avoiding the long, challenging, and technically-demanding process required for cryo-liftout. #1:  Journal: BioRxiv / Year: 2021 Journal: BioRxiv / Year: 2021Title: Waffle Method: A general and flexible approach for improving throughput in FIB-milling Authors: Kelley K / Raczkowski AM / Klykov O / Jaroenlak P / Bobe D / Kopylov M / Eng ET / Bhabha G / Potter CS / Carragher B / Noble AJ | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26396.map.gz emd_26396.map.gz | 8.5 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26396-v30.xml emd-26396-v30.xml emd-26396.xml emd-26396.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26396.png emd_26396.png | 210.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26396 http://ftp.pdbj.org/pub/emdb/structures/EMD-26396 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26396 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26396 | HTTPS FTP |
-Validation report
| Summary document |  emd_26396_validation.pdf.gz emd_26396_validation.pdf.gz | 280.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26396_full_validation.pdf.gz emd_26396_full_validation.pdf.gz | 280 KB | Display | |
| Data in XML |  emd_26396_validation.xml.gz emd_26396_validation.xml.gz | 4.8 KB | Display | |
| Data in CIF |  emd_26396_validation.cif.gz emd_26396_validation.cif.gz | 5.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26396 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26396 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26396 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26396 | HTTPS FTP |
-Related structure data
| EM raw data |  EMPIAR-10981 (Title: CryoET of E. coli prepared with the Waffle Method / Data size: 94.0 EMPIAR-10981 (Title: CryoET of E. coli prepared with the Waffle Method / Data size: 94.0 Data #1: Unaligned multi-frame micrographs for each tilt image [micrographs - multiframe]) |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26396.map.gz / Format: CCP4 / Size: 9.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26396.map.gz / Format: CCP4 / Size: 9.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E. coli waffle lamella tomogram from Appion-Protomo alignment. Pixelsize is actually 8.30616; ignore the filename. | ||||||||||||||||||||||||||||||||
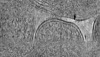
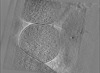
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 8.30616 Å | ||||||||||||||||||||||||||||||||
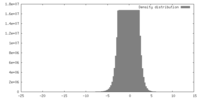
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : E. coli BL21 (DE3) cells
| Entire | Name: E. coli BL21 (DE3) cells |
|---|---|
| Components |
|
-Supramolecule #1: E. coli BL21 (DE3) cells
| Supramolecule | Name: E. coli BL21 (DE3) cells / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: PBS |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER |
| Vitrification | Cryogen name: NITROGEN / Details: HPF. |
| High pressure freezing | Instrument: OTHER Details: The value given for _emd_high_pressure_freezing.instrument is Wohlwend HPF Compact 01. This is not in a list of allowed values {'EMS-002 RAPID IMMERSION FREEZER', 'LEICA EM HPM100', 'OTHER', ...Details: The value given for _emd_high_pressure_freezing.instrument is Wohlwend HPF Compact 01. This is not in a list of allowed values {'EMS-002 RAPID IMMERSION FREEZER', 'LEICA EM HPM100', 'OTHER', 'LEICA EM PACT', 'BAL-TEC HPM 010', 'LEICA EM PACT2'} so OTHER is written into the XML file. |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 kV / Focused ion beam - Current: 3 nA / Focused ion beam - Duration: 3600 sec. / Focused ion beam - Temperature: 70 K / Focused ion beam - Initial thickness: 10000 nm / Focused ion beam - Final thickness: 200 nm Focused ion beam - Details: Waffle Method. The value given for _emd_sectioning_focused_ion_beam.instrument is TFS Aquilos 2. This is not in a list of allowed values {'OTHER', 'DB235'} so OTHER is ...Focused ion beam - Details: Waffle Method. The value given for _emd_sectioning_focused_ion_beam.instrument is TFS Aquilos 2. This is not in a list of allowed values {'OTHER', 'DB235'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 4.87 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 6.0 µm / Nominal defocus min: 6.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: SIMULTANEOUS ITERATIVE (SIRT) / Software - Name: TOMO3D (ver. Jan 2015) / Software - details: Dose weighted / Number images used: 36 |
|---|
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)