+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse retromer sub-structure: VPS35/VPS35 flat substructure | |||||||||
 Map data Map data | VPS35/VPS35 flat substructure | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Kendall AK / Jackson LP | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2022 Journal: J Biol Chem / Year: 2022Title: Improved mammalian retromer cryo-EM structures reveal a new assembly interface. Authors: Amy K Kendall / Mintu Chandra / Boyang Xie / William Wan / Lauren P Jackson /  Abstract: Retromer (VPS26/VPS35/VPS29 subunits) assembles with multiple sorting nexin proteins on membranes to mediate endosomal recycling of transmembrane protein cargoes. Retromer has been implicated in ...Retromer (VPS26/VPS35/VPS29 subunits) assembles with multiple sorting nexin proteins on membranes to mediate endosomal recycling of transmembrane protein cargoes. Retromer has been implicated in other cellular processes, including mitochondrial homeostasis, nutrient sensing, autophagy, and fission events. Mechanisms for mammalian retromer assembly remain undefined, and retromer engages multiple sorting nexin proteins to sort cargoes to different destinations. Published structures demonstrate mammalian retromer forms oligomers in vitro, but several structures were poorly resolved. We report here improved retromer oligomer structures using single-particle cryo-EM by combining data collected from tilted specimens with multiple advancements in data processing, including using a 3D starting model for enhanced automated particle picking in RELION. We used a retromer mutant (3KE retromer) that breaks VPS35-mediated interfaces to determine a structure of a new assembly interface formed by the VPS26A and VPS35 N-termini. The interface reveals how an N-terminal VPS26A arrestin saddle can link retromer chains by engaging a neighboring VPS35 N- terminus, on the opposite side from the well-characterized C-VPS26/N-VPS35 interaction observed within heterotrimers. The new interaction interface exhibits substantial buried surface area (∼7000 Å) and further suggests that metazoan retromer may serve as an adaptable scaffold. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26345.map.gz emd_26345.map.gz | 15.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26345-v30.xml emd-26345-v30.xml emd-26345.xml emd-26345.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26345.png emd_26345.png | 51.8 KB | ||
| Others |  emd_26345_half_map_1.map.gz emd_26345_half_map_1.map.gz emd_26345_half_map_2.map.gz emd_26345_half_map_2.map.gz | 12.8 MB 12.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26345 http://ftp.pdbj.org/pub/emdb/structures/EMD-26345 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26345 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26345 | HTTPS FTP |
-Validation report
| Summary document |  emd_26345_validation.pdf.gz emd_26345_validation.pdf.gz | 625.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26345_full_validation.pdf.gz emd_26345_full_validation.pdf.gz | 625.2 KB | Display | |
| Data in XML |  emd_26345_validation.xml.gz emd_26345_validation.xml.gz | 9.5 KB | Display | |
| Data in CIF |  emd_26345_validation.cif.gz emd_26345_validation.cif.gz | 11 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26345 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26345 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26345 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26345 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26345.map.gz / Format: CCP4 / Size: 16.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26345.map.gz / Format: CCP4 / Size: 16.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | VPS35/VPS35 flat substructure | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.096 Å | ||||||||||||||||||||||||||||||||||||
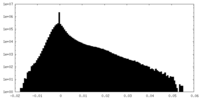
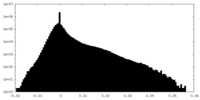
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: VPS35/VPS35 flat substructure, half map 1
| File | emd_26345_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | VPS35/VPS35 flat substructure, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
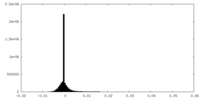
| Density Histograms |
-Half map: VPS35/VPS35 flat substructure, half map 2
| File | emd_26345_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | VPS35/VPS35 flat substructure, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
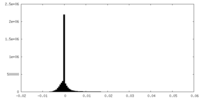
| Density Histograms |
- Sample components
Sample components
-Entire : Mouse retromer sub-structure: VPS35/VPS35 flat dimer
| Entire | Name: Mouse retromer sub-structure: VPS35/VPS35 flat dimer |
|---|---|
| Components |
|
-Supramolecule #1: Mouse retromer sub-structure: VPS35/VPS35 flat dimer
| Supramolecule | Name: Mouse retromer sub-structure: VPS35/VPS35 flat dimer / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #1: Vacuolar protein sorting-associated protein 35
| Macromolecule | Name: Vacuolar protein sorting-associated protein 35 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MPTTQQSPQD EQEKLLDEA I QAVKVQSF QM KRCLDKN KLM DALKHA SNML GELRT SMLSP KSYY ELYMA ISDE LHYLEV YLT DEFAKGR KV ADLYELVQ Y AGNIIPRLY LLITVGVVYV K SFPQSRK D ILKDLVEM CR GVQHPLR GLF LRNYLL QCTR ...String: MPTTQQSPQD EQEKLLDEA I QAVKVQSF QM KRCLDKN KLM DALKHA SNML GELRT SMLSP KSYY ELYMA ISDE LHYLEV YLT DEFAKGR KV ADLYELVQ Y AGNIIPRLY LLITVGVVYV K SFPQSRK D ILKDLVEM CR GVQHPLR GLF LRNYLL QCTR NILPD EGEPT DEET TGDISD SM DFVLLNF AE MNKLWVRM Q HQGHSRDRE KRERERQELR ILVGTNLVR L SQLEGVNV ER YKQ IVL TGI LEQVVN CRDA LAQEY LMECI IQVF PDEFHL QTL NPFLRAC AE LHQNVNVK N III ALIDR LALFAHREDG PGIPAEIKL F DIFSQQVA TV IQSRQDM PSE DVVSLQ VSLI NLAMK CYPD RVDY VDKVLE TTV EIFNKLN LE HIATSSAV S KELTRLLKI PVDTYNNILT VLKLKH FH P LFEYFDYE SR KSMSCYV LSN VLDYNT EIVS QDQVD SIMNL VSTL IQDQPD QPV EDPD PE DF ADEQSLVG R FIHLLRSDD PDQQYLILNT ARKHFGAGG N QRIRFTLP PL VFAAYQL A F RYKENS QMDD KWEKK CQKIF SFAH QTISAL IKA ELAELPL RL FLQGALAA G EIGFENHE T VAYEFMSQA FSLYEDEISD SKAQLAAIT L IIGTFERM KC FSEENHE PLR TQCALA ASKL LKKPD QGRA VSTCA HLFWS GRNT DKNGEE LHG GKRVMEC LK KALKIANQ C MDPSLQVQL F IEILNRYI YFYEKENDA V TIQVLNQL IQ KIREDLP NLE SSEETE QINK HFHNT LEHLR SR R ESPESE GPI YEGLIL |
-Macromolecule #2: Vacuolar protein sorting-associated protein 26A
| Macromolecule | Name: Vacuolar protein sorting-associated protein 26A / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSFLGGFFGP ICEIDVALN D GETRKMAE MK TEDGKVE KHY LFYDGE SVSG KVNLA FKQPG KRLE HQGIR IEFV GQIELF NDK SNTHEFV NL VKELALPG E LTQSRSYDF EFMQVEKPYE S YIGANVR L RYFLKVTI VR RLTDLVK EYD LIVHQL ATYP ...String: MSFLGGFFGP ICEIDVALN D GETRKMAE MK TEDGKVE KHY LFYDGE SVSG KVNLA FKQPG KRLE HQGIR IEFV GQIELF NDK SNTHEFV NL VKELALPG E LTQSRSYDF EFMQVEKPYE S YIGANVR L RYFLKVTI VR RLTDLVK EYD LIVHQL ATYP DVNNS IKMEV GIED CLHIEF EY NKSKYHL KD VIVGKIYF L LVRIKIQHM ELQLIKKEIT GIGPSTTTE T ETIAKYEI MD GAP VKG ESI PIRLFL AGYD PTPTM RDVNK KFSV RYFLNL VLV DEEDRRY FK QQEIILWR K APE KLRKQ RTNFHQRFES PDSQASAEQ P EM |
-Macromolecule #3: vacuolar protein sorting-associated protein 29
| Macromolecule | Name: vacuolar protein sorting-associated protein 29 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MLVLVLGDLH IPHRCNSLP A KFKKLLVP GK IQHILCT GNL CTKESY DYLK TLAGD VHIVR GDFD ENLNY PEQK VVTVGQ FKI GLIHGHQ VI PWGDMASL A LLQRQFDVD ILISGHTHKF E AFEHENK F YINPGSAT GA YNALET |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 72.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.7 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 69381 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)