[English] 日本語
 Yorodumi
Yorodumi- EMDB-26262: SARS-Cov2 S protein structure in complex with neutralizing monocl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
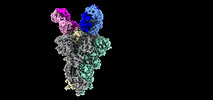
| Title | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | |||||||||
 Map data Map data | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-Cov2 6P spike protein / immune complex / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
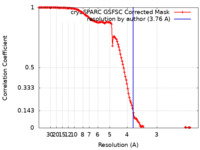
| Method | single particle reconstruction / cryo EM / Resolution: 3.76 Å | |||||||||
 Authors Authors | Patel A / Ortlund E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody. Authors: Sanjeev Kumar / Anamika Patel / Lilin Lai / Chennareddy Chakravarthy / Rajesh Valanparambil / Elluri Seetharami Reddy / Kamalvishnu Gottimukkala / Meredith E Davis-Gardner / Venkata ...Authors: Sanjeev Kumar / Anamika Patel / Lilin Lai / Chennareddy Chakravarthy / Rajesh Valanparambil / Elluri Seetharami Reddy / Kamalvishnu Gottimukkala / Meredith E Davis-Gardner / Venkata Viswanadh Edara / Susanne Linderman / Kaustuv Nayak / Kritika Dixit / Pragati Sharma / Prashant Bajpai / Vanshika Singh / Filipp Frank / Narayanaiah Cheedarla / Hans P Verkerke / Andrew S Neish / John D Roback / Grace Mantus / Pawan Kumar Goel / Manju Rahi / Carl W Davis / Jens Wrammert / Sucheta Godbole / Amy R Henry / Daniel C Douek / Mehul S Suthar / Rafi Ahmed / Eric Ortlund / Amit Sharma / Kaja Murali-Krishna / Anmol Chandele /   Abstract: In this study, by characterizing several human monoclonal antibodies (mAbs) isolated from single B cells of the COVID-19-recovered individuals in India who experienced ancestral Wuhan strain (WA.1) ...In this study, by characterizing several human monoclonal antibodies (mAbs) isolated from single B cells of the COVID-19-recovered individuals in India who experienced ancestral Wuhan strain (WA.1) of SARS-CoV-2 during early stages of the pandemic, we found a receptor binding domain (RBD)-specific mAb 002-S21F2 that has rare gene usage and potently neutralized live viral isolates of SARS-CoV-2 variants including Alpha, Beta, Gamma, Delta, and Omicron sublineages (BA.1, BA.2, BA.2.12.1, BA.4, and BA.5) with IC ranging from 0.02 to 0.13 μg/ml. Structural studies of 002-S21F2 in complex with spike trimers of Omicron and WA.1 showed that it targets a conformationally conserved epitope on the outer face of RBD (class 3 surface) outside the ACE2-binding motif, thereby providing a mechanistic insights for its broad neutralization activity. The discovery of 002-S21F2 and the broadly neutralizing epitope it targets have timely implications for developing a broad range of therapeutic and vaccine interventions against SARS-CoV-2 variants including Omicron sublineages. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26262.map.gz emd_26262.map.gz | 230.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26262-v30.xml emd-26262-v30.xml emd-26262.xml emd-26262.xml | 31 KB 31 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26262_fsc.xml emd_26262_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_26262.png emd_26262.png | 31.5 KB | ||
| Masks |  emd_26262_msk_1.map emd_26262_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26262.cif.gz emd-26262.cif.gz | 8.2 KB | ||
| Others |  emd_26262_additional_1.map.gz emd_26262_additional_1.map.gz emd_26262_additional_2.map.gz emd_26262_additional_2.map.gz emd_26262_additional_3.map.gz emd_26262_additional_3.map.gz emd_26262_half_map_1.map.gz emd_26262_half_map_1.map.gz emd_26262_half_map_2.map.gz emd_26262_half_map_2.map.gz | 227.4 MB 229.9 MB 229.9 MB 226.7 MB 226.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26262 http://ftp.pdbj.org/pub/emdb/structures/EMD-26262 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26262 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26262 | HTTPS FTP |
-Validation report
| Summary document |  emd_26262_validation.pdf.gz emd_26262_validation.pdf.gz | 866.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26262_full_validation.pdf.gz emd_26262_full_validation.pdf.gz | 865.7 KB | Display | |
| Data in XML |  emd_26262_validation.xml.gz emd_26262_validation.xml.gz | 21.5 KB | Display | |
| Data in CIF |  emd_26262_validation.cif.gz emd_26262_validation.cif.gz | 28 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26262 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26262 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26262 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26262 | HTTPS FTP |
-Related structure data
| Related structure data |  7u0pMC  7uplC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26262.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26262.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26262_msk_1.map emd_26262_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
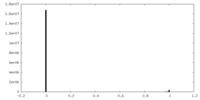
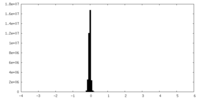
| Density Histograms |
-Additional map: Combine focused map
| File | emd_26262_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Combine focused map | ||||||||||||
| Projections & Slices |
| ||||||||||||
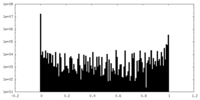
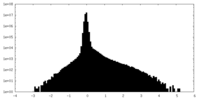
| Density Histograms |
-Additional map: RBD chainA mAb localrefine map
| File | emd_26262_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RBD_chainA mAb localrefine map | ||||||||||||
| Projections & Slices |
| ||||||||||||
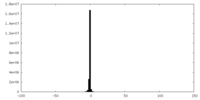
| Density Histograms |
-Additional map: RBD chainC mAblocalrefine map
| File | emd_26262_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RBD_chainC mAblocalrefine map | ||||||||||||
| Projections & Slices |
| ||||||||||||
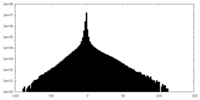
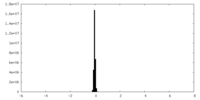
| Density Histograms |
-Half map: half map2
| File | emd_26262_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
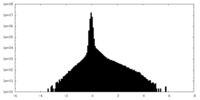
| Density Histograms |
-Half map: half map 1
| File | emd_26262_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-Cov2 Spike trimer in complex with monoclonal antibody
| Entire | Name: SARS-Cov2 Spike trimer in complex with monoclonal antibody |
|---|---|
| Components |
|
-Supramolecule #1: SARS-Cov2 Spike trimer in complex with monoclonal antibody
| Supramolecule | Name: SARS-Cov2 Spike trimer in complex with monoclonal antibody type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: Spike glycoprotein
| Supramolecule | Name: Spike glycoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: mAb 002-S21F2
| Supramolecule | Name: mAb 002-S21F2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 133.781312 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSPG SASSVASQSI IAYTMSLGAE NSVAYSNNSI AIPTNFTI S VTTEILPVSM TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGF NFSQILPDPS KPSKRSPIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFG AGPALQIPFP MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STPSALGKLQ DVVNQNAQAL N TLVKQLSS NFGAISSVLN DILSRLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RV DFCGKGY HLMSFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNT FVSGNC DVVIGIVNNT VYDPLQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDL QELGK YEQ UniProtKB: Spike glycoprotein |
-Macromolecule #2: mAb 002-S21F2 Heavy chain
| Macromolecule | Name: mAb 002-S21F2 Heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.323574 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVQSGAE VKKPGESLKI SCKASGYNFA YYWIGWVRQM PGKGLEWMGI IYPGDSDTRY SPSFQGQVTI SADKSISTAY LQWSSLKAS DTAMYYCARG EMTAVFGDYW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS W NSGALTSG ...String: EVQLVQSGAE VKKPGESLKI SCKASGYNFA YYWIGWVRQM PGKGLEWMGI IYPGDSDTRY SPSFQGQVTI SADKSISTAY LQWSSLKAS DTAMYYCARG EMTAVFGDYW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS W NSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKRVE PKSCDKTHTC PPCPAPELLG GP SVFLFPP KPKDTLMISR TPEVTCVVVD VSHEDPEVKF NWYVDGVEVH NAKTKPREEQ YNSTYRVVSV LTVLHQDWLN GKE YKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVL DSDGS FFLYSKLTVD KSRWQQGNVF SCSVMHEALH NHYTQKSLSL SPGK |
-Macromolecule #3: mAb 002_S21F2 light chain
| Macromolecule | Name: mAb 002_S21F2 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.648248 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCQASQDIS NYLNWYQQKP GKAPKLLIYD ASNLETGVPS RFSGSGSGTD FTLTISSLQP EDIATYYCQ QYKILLTWTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: DIQMTQSPSS LSASVGDRVT ITCQASQDIS NYLNWYQQKP GKAPKLLIYD ASNLETGVPS RFSGSGSGTD FTLTISSLQP EDIATYYCQ QYKILLTWTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 22 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 297 K / Instrument: FEI VITROBOT MARK IV / Details: Wait time 20 seconds and blot time 3 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 2 / Number real images: 3863 / Average electron dose: 51.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 79000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-7u0p: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)