[English] 日本語
 Yorodumi
Yorodumi- EMDB-26259: State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall map -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall map | ||||||||||||
 Map data Map data | Map of state NE1 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ribosome biogenesis / DEAD-box ATPases / methyltransferase / nucleolus / RIBOSOME | ||||||||||||
| Function / homology |  Function and homology information Function and homology information27S pre-rRNA (guanosine2922-2'-O)-methyltransferase / rRNA (guanosine-2'-O-)-methyltransferase activity / exonucleolytic trimming to generate mature 5'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA (uridine-2'-O-)-methyltransferase activity / rRNA (guanine) methyltransferase activity / nuclear exosome (RNase complex) / PeBoW complex / rRNA primary transcript binding / positive regulation of ATP-dependent activity / maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) ...27S pre-rRNA (guanosine2922-2'-O)-methyltransferase / rRNA (guanosine-2'-O-)-methyltransferase activity / exonucleolytic trimming to generate mature 5'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA (uridine-2'-O-)-methyltransferase activity / rRNA (guanine) methyltransferase activity / nuclear exosome (RNase complex) / PeBoW complex / rRNA primary transcript binding / positive regulation of ATP-dependent activity / maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA methylation / maturation of 5.8S rRNA / hexon binding / pre-mRNA 5'-splice site binding / cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / proteasome binding / Major pathway of rRNA processing in the nucleolus and cytosol / SRP-dependent cotranslational protein targeting to membrane / GTP hydrolysis and joining of the 60S ribosomal subunit / ribosomal large subunit binding / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Formation of a pool of free 40S subunits / negative regulation of mRNA splicing, via spliceosome / preribosome, large subunit precursor / nuclear-transcribed mRNA catabolic process / ATPase activator activity / L13a-mediated translational silencing of Ceruloplasmin expression / translational elongation / ribosomal large subunit export from nucleus / regulation of translational fidelity / protein-RNA complex assembly / ribonucleoprotein complex binding / ribosomal subunit export from nucleus / maturation of LSU-rRNA / translation initiation factor activity / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / proteasome complex / assembly of large subunit precursor of preribosome / ribosomal large subunit biogenesis / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / cytosolic ribosome assembly / small-subunit processome / macroautophagy / maintenance of translational fidelity / protein catabolic process / rRNA processing / protein transport / ribosome biogenesis / viral capsid / protein-macromolecule adaptor activity / ATPase binding / 5S rRNA binding / large ribosomal subunit rRNA binding / ribosomal large subunit assembly / cytoplasmic translation / cytosolic large ribosomal subunit / negative regulation of translation / rRNA binding / ribosome / structural constituent of ribosome / ribonucleoprotein complex / translation / GTPase activity / mRNA binding / host cell nucleus / GTP binding / nucleolus / RNA binding / nucleoplasm / identical protein binding / nucleus / metal ion binding / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
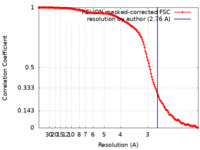
| Method | single particle reconstruction / cryo EM / Resolution: 2.76 Å | ||||||||||||
 Authors Authors | Cruz VE / Sekulski K / Peddada N / Erzberger JP | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Sequence-specific remodeling of a topologically complex RNP substrate by Spb4. Authors: Victor Emmanuel Cruz / Kamil Sekulski / Nagesh Peddada / Carolin Sailer / Sahana Balasubramanian / Christine S Weirich / Florian Stengel / Jan P Erzberger /    Abstract: DEAD-box ATPases are ubiquitous enzymes essential in all aspects of RNA biology. However, the limited in vitro catalytic activities described for these enzymes are at odds with their complex cellular ...DEAD-box ATPases are ubiquitous enzymes essential in all aspects of RNA biology. However, the limited in vitro catalytic activities described for these enzymes are at odds with their complex cellular roles, most notably in driving large-scale RNA remodeling steps during the assembly of ribonucleoproteins (RNPs). We describe cryo-EM structures of 60S ribosomal biogenesis intermediates that reveal how context-specific RNA unwinding by the DEAD-box ATPase Spb4 results in extensive, sequence-specific remodeling of rRNA secondary structure. Multiple cis and trans interactions stabilize Spb4 in a post-catalytic, high-energy intermediate that drives the organization of the three-way junction at the base of rRNA domain IV. This mechanism explains how limited strand separation by DEAD-box ATPases is leveraged to provide non-equilibrium directionality and ensure efficient and accurate RNP assembly. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26259.map.gz emd_26259.map.gz | 229 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26259-v30.xml emd-26259-v30.xml emd-26259.xml emd-26259.xml | 77.4 KB 77.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26259_fsc.xml emd_26259_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_26259.png emd_26259.png | 99 KB | ||
| Filedesc metadata |  emd-26259.cif.gz emd-26259.cif.gz | 16.3 KB | ||
| Others |  emd_26259_half_map_1.map.gz emd_26259_half_map_1.map.gz emd_26259_half_map_2.map.gz emd_26259_half_map_2.map.gz | 194.1 MB 193.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26259 http://ftp.pdbj.org/pub/emdb/structures/EMD-26259 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26259 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26259 | HTTPS FTP |
-Validation report
| Summary document |  emd_26259_validation.pdf.gz emd_26259_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26259_full_validation.pdf.gz emd_26259_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_26259_validation.xml.gz emd_26259_validation.xml.gz | 21.8 KB | Display | |
| Data in CIF |  emd_26259_validation.cif.gz emd_26259_validation.cif.gz | 28.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26259 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26259 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26259 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26259 | HTTPS FTP |
-Related structure data
| Related structure data |  7u0hMC  7nacC  7nadC  7nafC  7r6kC  7r6qC  7r72C  7r7aC  7r7cC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26259.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26259.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of state NE1 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02885 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-map 2 of state NE1
| File | emd_26259_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 of state NE1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1 of state NE1
| File | emd_26259_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 of state NE1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Early nucleoplasmic 60S intermediate purified with tags on Nop53 ...
+Supramolecule #1: Early nucleoplasmic 60S intermediate purified with tags on Nop53 ...
+Macromolecule #1: 25S rRNA
+Macromolecule #2: 5.8S rRNA
+Macromolecule #3: ITS2 rRNA
+Macromolecule #4: 60S ribosomal protein L2-A
+Macromolecule #5: 60S ribosomal protein L3
+Macromolecule #6: 60S ribosomal protein L4-A
+Macromolecule #7: 60S ribosomal protein L6-A
+Macromolecule #8: 60S ribosomal protein L7-A
+Macromolecule #9: 60S ribosomal protein L8-A
+Macromolecule #10: 60S ribosomal protein L9-A
+Macromolecule #11: Proteasome-interacting protein CIC1
+Macromolecule #12: 60S ribosomal protein L13-A
+Macromolecule #13: 60S ribosomal protein L14-A
+Macromolecule #14: 60S ribosomal protein L15-A
+Macromolecule #15: 60S ribosomal protein L16-A
+Macromolecule #16: 60S ribosomal protein L17-A
+Macromolecule #17: 60S ribosomal protein L18-A
+Macromolecule #18: 60S ribosomal protein L19-A
+Macromolecule #19: 60S ribosomal protein L20-A
+Macromolecule #20: 60S ribosomal protein L21-A
+Macromolecule #21: 60S ribosomal protein L22-A
+Macromolecule #22: 60S ribosomal protein L23-A
+Macromolecule #23: Ribosome assembly factor MRT4
+Macromolecule #24: 60S ribosomal protein L25
+Macromolecule #25: 60S ribosomal protein L26-A
+Macromolecule #26: 60S ribosomal protein L27-A
+Macromolecule #27: 60S ribosomal protein L28
+Macromolecule #28: Nucleolar GTP-binding protein 1
+Macromolecule #29: 60S ribosomal protein L30
+Macromolecule #30: 60S ribosomal protein L31-A
+Macromolecule #31: 60S ribosomal protein L32
+Macromolecule #32: 60S ribosomal protein L33-A
+Macromolecule #33: 60S ribosomal protein L34-A
+Macromolecule #34: 60S ribosomal protein L35-A
+Macromolecule #35: 60S ribosomal protein L36-A
+Macromolecule #36: 60S ribosomal protein L37-A
+Macromolecule #37: 60S ribosomal protein L38
+Macromolecule #38: Nucleolar GTP-binding protein 2
+Macromolecule #39: Pescadillo homolog
+Macromolecule #40: Ribosome biogenesis protein 15
+Macromolecule #41: 60S ribosomal protein L43-A
+Macromolecule #42: Ribosome biogenesis protein NOP53
+Macromolecule #43: Ribosome biogenesis protein NSA2
+Macromolecule #44: Nuclear GTP-binding protein NUG1
+Macromolecule #45: Ribosome biogenesis protein RLP7
+Macromolecule #46: Ribosome biogenesis protein RLP24
+Macromolecule #47: 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase
+Macromolecule #48: Eukaryotic translation initiation factor 6
+Macromolecule #49: UPF0642 protein YBL028C
+Macromolecule #50: MAGNESIUM ION
+Macromolecule #51: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.3 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 7398 / Average exposure time: 0.05 sec. / Average electron dose: 1.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7u0h: |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)