+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
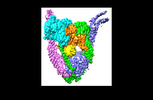
| Title | ATP and DNA bound SMC5/6 core complex | |||||||||
 Map data Map data | Primary map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nse1 / Nse3 / Nse4 / Smc5 / Smc6 / Nse2 / ATP / DNA BINDING PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationSmc5-Smc6 complex / resolution of DNA recombination intermediates / DNA double-strand break attachment to nuclear envelope / chromosome separation / SUMOylation of DNA damage response and repair proteins / Platelet degranulation / chromatin looping / recombinational repair / regulation of telomere maintenance / protein sumoylation ...Smc5-Smc6 complex / resolution of DNA recombination intermediates / DNA double-strand break attachment to nuclear envelope / chromosome separation / SUMOylation of DNA damage response and repair proteins / Platelet degranulation / chromatin looping / recombinational repair / regulation of telomere maintenance / protein sumoylation / double-strand break repair via homologous recombination / site of double-strand break / single-stranded DNA binding / chromosome, telomeric region / damaged DNA binding / DNA repair / negative regulation of transcription by RNA polymerase II / ATP hydrolysis activity / mitochondrion / ATP binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Yu Y / Patel DJ | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Cryo-EM structure of DNA-bound Smc5/6 reveals DNA clamping enabled by multi-subunit conformational changes. Authors: You Yu / Shibai Li / Zheng Ser / Huihui Kuang / Thane Than / Danying Guan / Xiaolan Zhao / Dinshaw J Patel /   Abstract: Structural maintenance of chromosomes (SMC) complexes are essential for chromatin organization and functions throughout the cell cycle. The cohesin and condensin SMCs fold and tether DNA, while ...Structural maintenance of chromosomes (SMC) complexes are essential for chromatin organization and functions throughout the cell cycle. The cohesin and condensin SMCs fold and tether DNA, while Smc5/6 directly promotes DNA replication and repair. The functions of SMCs rely on their abilities to engage DNA, but how Smc5/6 binds and translocates on DNA remains largely unknown. Here, we present a 3.8 Å cryogenic electron microscopy (cryo-EM) structure of DNA-bound Saccharomyces cerevisiae Smc5/6 complex containing five of its core subunits, including Smc5, Smc6, and the Nse1-3-4 subcomplex. Intricate interactions among these subunits support the formation of a clamp that encircles the DNA double helix. The positively charged inner surface of the clamp contacts DNA in a nonsequence-specific manner involving numerous DNA binding residues from four subunits. The DNA duplex is held up by Smc5 and 6 head regions and positioned between their coiled-coil arm regions, reflecting an engaged-head and open-arm configuration. The Nse3 subunit secures the DNA from above, while the hook-shaped Nse4 kleisin forms a scaffold connecting DNA and all other subunits. The Smc5/6 DNA clamp shares similarities with DNA-clamps formed by other SMCs but also exhibits differences that reflect its unique functions. Mapping cross-linking mass spectrometry data derived from DNA-free Smc5/6 to the DNA-bound Smc5/6 structure identifies multi-subunit conformational changes that enable DNA capture. Finally, mutational data from cells reveal distinct DNA binding contributions from each subunit to Smc5/6 chromatin association and cell fitness. In summary, our integrative study illuminates how a unique SMC complex engages DNA in supporting genome regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26140.map.gz emd_26140.map.gz | 6.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26140-v30.xml emd-26140-v30.xml emd-26140.xml emd-26140.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26140.png emd_26140.png | 50.9 KB | ||
| Filedesc metadata |  emd-26140.cif.gz emd-26140.cif.gz | 8.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26140 http://ftp.pdbj.org/pub/emdb/structures/EMD-26140 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26140 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26140 | HTTPS FTP |
-Validation report
| Summary document |  emd_26140_validation.pdf.gz emd_26140_validation.pdf.gz | 344.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26140_full_validation.pdf.gz emd_26140_full_validation.pdf.gz | 343.9 KB | Display | |
| Data in XML |  emd_26140_validation.xml.gz emd_26140_validation.xml.gz | 6.1 KB | Display | |
| Data in CIF |  emd_26140_validation.cif.gz emd_26140_validation.cif.gz | 7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26140 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26140 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26140 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26140 | HTTPS FTP |
-Related structure data
| Related structure data |  7tveMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26140.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26140.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.069 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Hexamer complex of Smc5-Smc6-Nse1-Nse3-Nse4-Nse2 with ATP and DNA
| Entire | Name: Hexamer complex of Smc5-Smc6-Nse1-Nse3-Nse4-Nse2 with ATP and DNA |
|---|---|
| Components |
|
-Supramolecule #1: Hexamer complex of Smc5-Smc6-Nse1-Nse3-Nse4-Nse2 with ATP and DNA
| Supramolecule | Name: Hexamer complex of Smc5-Smc6-Nse1-Nse3-Nse4-Nse2 with ATP and DNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 / Details: Smc5 E1015Q mutation and Smc6 E1048Q mutation |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 410 KDa |
-Macromolecule #1: DNA (68-MER)
| Macromolecule | Name: DNA (68-MER) / type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 21.253111 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA) (DA)(DA)(DA)(DA)(DA)(DA) ...String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA) |
-Macromolecule #2: DNA (78-MER)
| Macromolecule | Name: DNA (78-MER) / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.682055 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT) (DT)(DT)(DT)(DT)(DT)(DT) ...String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT) |
-Macromolecule #3: Non-structural maintenance of chromosomes element 1
| Macromolecule | Name: Non-structural maintenance of chromosomes element 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.437395 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEVHEEHVSA PVTGDATAKY LLQYILSARG ICHENALILA LMRLETDAST LNTEWSIQQW VDKLNDYINA INVKLNLLGY KIIRINHGI GRNAVTLKAK QNFESFEDNT AIRAHDNDYA VLQSIVLPES NRFFVYVNLA STEETKLATR FNQNEIEFIK W AIEQFMIS ...String: MEVHEEHVSA PVTGDATAKY LLQYILSARG ICHENALILA LMRLETDAST LNTEWSIQQW VDKLNDYINA INVKLNLLGY KIIRINHGI GRNAVTLKAK QNFESFEDNT AIRAHDNDYA VLQSIVLPES NRFFVYVNLA STEETKLATR FNQNEIEFIK W AIEQFMIS GETIVEGPAL DTSIIVREVN RILVAATGDS NLAKWRKFST FTVGSTNLFQ FQELTATDIE DLLLRLCELK WF YRTQEGK FGIDLRCIAE LEEYLTSMYN LNTCQNCHKL AIQGVRCGNE SCREENEETG ENSLSQIWHV DCFKHYITHV SKN CDRCGS SLITEGVYVI G UniProtKB: UNIPROTKB: A0A7I9FFW3 |
-Macromolecule #4: Structural maintenance of chromosomes protein 6
| Macromolecule | Name: Structural maintenance of chromosomes protein 6 / type: protein_or_peptide / ID: 4 / Details: Smc6(E1048Q)-2XStrep / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 132.445078 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MISTTISGKR PIEQVDDELL SLTAQQENEE QQQQRKRRRH QFAPMTQFNS NTLDEDSGFR SSSDVATADQ DNFLEESPSG YIKKVILRN FMCHEHFELE LGSRLNFIVG NNGSGKSAIL TAITIGLGAK ASETNRGSSL KDLIREGCYS AKIILHLDNS K YGAYQQGI ...String: MISTTISGKR PIEQVDDELL SLTAQQENEE QQQQRKRRRH QFAPMTQFNS NTLDEDSGFR SSSDVATADQ DNFLEESPSG YIKKVILRN FMCHEHFELE LGSRLNFIVG NNGSGKSAIL TAITIGLGAK ASETNRGSSL KDLIREGCYS AKIILHLDNS K YGAYQQGI FGNEIIVERI IKRDGPASFS LRSENGKEIS NKKKDIQTVV DYFSVPVSNP MCFLSQDAAR SFLTASTSQD KY SHFMKGT LLQEITENLL YASAIHDSAQ ENMALHLENL KSLKAEYEDA KKLLRELNQT SDLNERKMLL QAKSLWIDVA HNT DACKNL ENEISGIQQK VDEVTEKIRN RQEKIERYTS DGTTIEAQID AKVIYVNEKD SEHQNARELL RDVKSRFEKE KSNQ AEAQS NIDQGRKKVD ALNKTIAHLE EELTKEMGGD KDQMRQELEQ LEKANEKLRE VNNSLVVSLQ DVKNEERDIQ HERES ELRT ISRSIQNKKV ELQNIAKGND TFLMNFDRNM DRLLRTIEQR KNEFETPAIG PLGSLVTIRK GFEKWTRSIQ RAISSS LNA FVVSNPKDNR LFRDIMRSCG IRSNIPIVTY CLSQFDYSKG RAHGNYPTIV DALEFSKPEI ECLFVDLSRI ERIVLIE DK NEARNFLQRN PVNVNMALSL RDRRSGFQLS GGYRLDTVTY QDKIRLKVNS SSDNGTQYLK DLIEQETKEL QNIRDRYE E KLSEVRSRLK EIDGRLKSTK NEMRKTNFRM TELKMNVGKV VDTGILNSKI NERKNQEQAI ASYEAAKEEL GLKIEQIAQ EAQPIKEQYD STKLALVEAQ DELQQLKEDI NSRQSKIQKY KDDTIYYEDK KKVYLENIKK IEVNVAALKE GIQRQIQNAC AFCSKERIE NVDLPDTQEE IKRELDKVSR MIQKAEKSLG LSQEEVIALF EKCRNKYKEG QKKYMEIDEA LNRLHNSLKA R DQNYKNAE KGTCFDADMD FRASLKVRKF SGNLSFIKDT KSLEIYILTT NDEKARNVDT LSGGEKSFSQ MALLLATWKP MR SRIIALD QFDVFMDQVN RKIGTTLIVK KLKDIARTQT IIITPQDIGK IADIDSSGVS IHRMRDPERQ NNSNFYNGSL EVL FQGPGG SSAWSHPQFE KGGGSGGGSG GGSWSHPQFE K UniProtKB: Structural maintenance of chromosomes protein 6 |
-Macromolecule #5: Structural maintenance of chromosomes protein 5
| Macromolecule | Name: Structural maintenance of chromosomes protein 5 / type: protein_or_peptide / ID: 5 / Details: Smc5 (E1015Q) / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 126.293234 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTSLIDLGRY VERTHHGEDT EPRSKRVKIA KPDLSSFQPG SIIKIRLQDF VTYTLTEFNL SPSLNMIIGP NGSGKSTFVC AVCLGLAGK PEYIGRSKKV EDFIKNGQDV SKIEITLKNS PNVTDIEYID ARDETIKITR IITRSKRRSD YLINDYQVSE S VVKTLVAQ ...String: MTSLIDLGRY VERTHHGEDT EPRSKRVKIA KPDLSSFQPG SIIKIRLQDF VTYTLTEFNL SPSLNMIIGP NGSGKSTFVC AVCLGLAGK PEYIGRSKKV EDFIKNGQDV SKIEITLKNS PNVTDIEYID ARDETIKITR IITRSKRRSD YLINDYQVSE S VVKTLVAQ LNIQLDNLCQ FLSQERVEEF ARLKSVKLLV ETIRSIDASL LDVLDELREL QGNEQSLQKD LDFKKAKIVH LR QESDKLR KSVESLRDFQ NKKGEIELHS QLLPYVKVKD HKEKLNIYKE EYERAKANLR AILKDKKPFA NTKKTLENQV EEL TEKCSL KTDEFLKAKE KINEIFEKLN TIRDEVIKKK NQNEYYRGRT KKLQATIIST KEDFLRSQEI LAQTHLPEKS VFED IDIKR KEIINKEGEI RDLISEIDAK ANAINHEMRS IQRQAESKTK SLTTTDKIGI LNQDQDLKEV RDAVLMVREH PEMKD KILE PPIMTVSAIN AQFAAYLAQC VDYNTSKALT VVDSDSYKLF ANPILDKFKV NLRELSSADT TPPVPAETVR DLGFEG YLS DFITGDKRVM KMLCQTSKIH TIPVSRRELT PAQIKKLITP RPNGKILFKR IIHGNRLVDI KQSAYGSKQV FPTDVSI KQ TNFYQGSIMS NEQKIRIENE IINLKNEYND RKSTLDALSN QKSGYRHELS ELASKNDDIN REAHQLNEIR KKYTMRKS T IETLREKLDQ LKREARKDVS QKIKDIDDQI QQLLLKQRHL LSKMASSMKS LKNCQKELIS TQILQFEAQN MDVSMNDVI GFFNEREADL KSQYEDKKKF VKEMRDTPEF QSWMREIRSY DQDTKEKLNK VAEKYEEEGN FNLSFVQDVL DKLESEIAMV NHDESAVTI LDQVTAELRE LEHTVPQQSK DLETIKAKLK EDHAVLEPKL DDIVSKISAR FARLFNNVGS AGAVRLEKPK D YAEWKIEI MVKFRDNAPL KKLDSHTQSG GERAVSTVLY MIALQEFTSA PFRVVDQINQ GMDSRNERIV HKAMVENACA EN TSQYFLI TPKLLTGLHY HEKMRIHCVM AGSWIPNPSE DPKMIHFGET SNYSFDG UniProtKB: Structural maintenance of chromosomes protein 5 |
-Macromolecule #6: Non-structural maintenance of chromosome element 3
| Macromolecule | Name: Non-structural maintenance of chromosome element 3 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.193777 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMSSIDNDSD VDLTEDLAVA KIVKENPVAR KMVRYILSRG ESQNSIITRN KLQSVIHEAA REENIAKPSF SKMFMDINAI LYNVYGFEL QGLPSKNNMN AGGNGSNSNT NKSMPEPLGH RAQKFILLNN VPHSKNFDDF KILQSAHTYE ELIVTGEYIG D DIASGTSN ...String: MMSSIDNDSD VDLTEDLAVA KIVKENPVAR KMVRYILSRG ESQNSIITRN KLQSVIHEAA REENIAKPSF SKMFMDINAI LYNVYGFEL QGLPSKNNMN AGGNGSNSNT NKSMPEPLGH RAQKFILLNN VPHSKNFDDF KILQSAHTYE ELIVTGEYIG D DIASGTSN TLESKLSTDR DLVYKGVLSV ILCIVFFSKN NILHQELIKF LETFGIPSDG SKIAILNITI EDLIKSLEKR EY IVRLEEK SDTDGEVISY RIGRRTQAEL GLESLEKLVQ EIMGLEKEQT KSLHDDIIKS IGDSYSIG UniProtKB: Non-structural maintenance of chromosome element 3 |
-Macromolecule #7: Non-structural maintenance of chromosome element 4
| Macromolecule | Name: Non-structural maintenance of chromosome element 4 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 46.252996 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSTVISRKR RNSTVTEPDS SGETRKQKKS RSDEKSSSSK DGDPQLEFKV LQGYRDLESE MHKGRAQVTR TGDIGVAMDN LNAVDSLFN KVIGIKNNGL FAHDARAMVS ISELAQISVR NLKFDDSRSM VNLENIVNSL KRYMLKEHFK LNNIAENRND L TLAADEQS ...String: MSSTVISRKR RNSTVTEPDS SGETRKQKKS RSDEKSSSSK DGDPQLEFKV LQGYRDLESE MHKGRAQVTR TGDIGVAMDN LNAVDSLFN KVIGIKNNGL FAHDARAMVS ISELAQISVR NLKFDDSRSM VNLENIVNSL KRYMLKEHFK LNNIAENRND L TLAADEQS AADQQEESDG DIDRTPDDNH TDKATSSFKA TSMRHSYLQQ FSHYNEFSQF NWFRIGALYN TISKNAPITD HL MGPLSIE KKPRVLTQRR RNNDQVGEKI TAEKITQHSL NSTQQETTPE QVKKCFKKLS KKLGPEGSIN LFKFIIDPNS FSR SIENLF YTSFLIKEGK LLMEHDEEGL PTIKIKQSIS HTDSRSKEIE RQRRRAAHQN HIIFQMDMPT WRKLIKKYNI TSPF LDG UniProtKB: Non-structural maintenance of chromosome element 4 |
-Macromolecule #8: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 8 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 53.55 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller