+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AtTPC1 D454N-EDTA state II | |||||||||
 Map data Map data | locally sharpened map using Deepemhancer | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of jasmonic acid biosynthetic process / seed germination / regulation of stomatal movement / plant-type vacuole / vacuole / vacuolar membrane / monoatomic ion channel complex / voltage-gated calcium channel activity / calcium-mediated signaling / calcium ion transport ...regulation of jasmonic acid biosynthetic process / seed germination / regulation of stomatal movement / plant-type vacuole / vacuole / vacuolar membrane / monoatomic ion channel complex / voltage-gated calcium channel activity / calcium-mediated signaling / calcium ion transport / calcium ion binding / Golgi apparatus / identical protein binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Dickinson MS / Stroud RM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Molecular basis of multistep voltage activation in plant two-pore channel 1. Authors: Miles Sasha Dickinson / Jinping Lu / Meghna Gupta / Irene Marten / Rainer Hedrich / Robert M Stroud /   Abstract: Voltage-gated ion channels confer excitability to biological membranes, initiating and propagating electrical signals across large distances on short timescales. Membrane excitation requires channels ...Voltage-gated ion channels confer excitability to biological membranes, initiating and propagating electrical signals across large distances on short timescales. Membrane excitation requires channels that respond to changes in electric field and couple the transmembrane voltage to gating of a central pore. To address the mechanism of this process in a voltage-gated ion channel, we determined structures of the plant two-pore channel 1 at different stages along its activation coordinate. These high-resolution structures of activation intermediates, when compared with the resting-state structure, portray a mechanism in which the voltage-sensing domain undergoes dilation and in-membrane plane rotation about the gating charge-bearing helix, followed by charge translocation across the charge transfer seal. These structures, in concert with patch-clamp electrophysiology, show that residues in the pore mouth sense inhibitory Ca and are allosterically coupled to the voltage sensor. These conformational changes provide insight into the mechanism of voltage-sensor domain activation in which activation occurs vectorially over a series of elementary steps. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25825.map.gz emd_25825.map.gz | 267.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25825-v30.xml emd-25825-v30.xml emd-25825.xml emd-25825.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25825.png emd_25825.png | 65.2 KB | ||
| Others |  emd_25825_additional_1.map.gz emd_25825_additional_1.map.gz emd_25825_half_map_1.map.gz emd_25825_half_map_1.map.gz emd_25825_half_map_2.map.gz emd_25825_half_map_2.map.gz | 290.2 MB 284.5 MB 284.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25825 http://ftp.pdbj.org/pub/emdb/structures/EMD-25825 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25825 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25825 | HTTPS FTP |
-Validation report
| Summary document |  emd_25825_validation.pdf.gz emd_25825_validation.pdf.gz | 525.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25825_full_validation.pdf.gz emd_25825_full_validation.pdf.gz | 525.2 KB | Display | |
| Data in XML |  emd_25825_validation.xml.gz emd_25825_validation.xml.gz | 16.8 KB | Display | |
| Data in CIF |  emd_25825_validation.cif.gz emd_25825_validation.cif.gz | 20 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25825 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25825 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25825 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25825 | HTTPS FTP |
-Related structure data
| Related structure data |  7tddMC  7tbgC  7tdeC  7tdfC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25825.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25825.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | locally sharpened map using Deepemhancer | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Sharpened map
| File | emd_25825_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
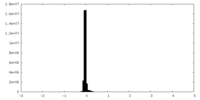
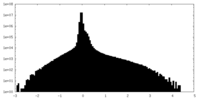
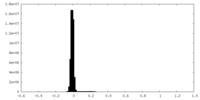
| Density Histograms |
-Half map: half map A
| File | emd_25825_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
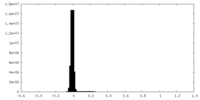
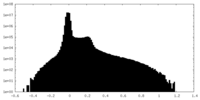
| Density Histograms |
-Half map: half map B
| File | emd_25825_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
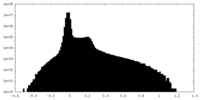
| Density Histograms |
- Sample components
Sample components
-Entire : AtTPC1 D454N-EDTA state II
| Entire | Name: AtTPC1 D454N-EDTA state II |
|---|---|
| Components |
|
-Supramolecule #1: AtTPC1 D454N-EDTA state II
| Supramolecule | Name: AtTPC1 D454N-EDTA state II / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 168 KDa |
-Macromolecule #1: Two pore calcium channel protein 1
| Macromolecule | Name: Two pore calcium channel protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 78.589844 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TDRVRRSEAI THGTPFQKAA ALVDLAEDGI GLPVEILDQS SFGESARYYF IFTRLDLIWS LNYFALLFLN FFEQPLWCEK NPKPSCKDR DYYYLGELPY LTNAESIIYE VITLAILLVH TFFPISYEGS RIFWTSRLNL VKVACVVILF VDVLVDFLYL S PLAFDFLP ...String: TDRVRRSEAI THGTPFQKAA ALVDLAEDGI GLPVEILDQS SFGESARYYF IFTRLDLIWS LNYFALLFLN FFEQPLWCEK NPKPSCKDR DYYYLGELPY LTNAESIIYE VITLAILLVH TFFPISYEGS RIFWTSRLNL VKVACVVILF VDVLVDFLYL S PLAFDFLP FRIAPYVRVI IFILSIRELR DTLVLLSGML GTYLNILALW MLFLLFASWI AFVMFEDTQQ GLTVFTSYGA TL YQMFILF TTSNNPDVWI PAYKSSRWSS VFFVLYVLIG VYFVTNLILA VVYDSFKEQL AKQVSGMDQM KRRMLEKAFG LID SDKNGE IDKNQCIKLF EQLTNYRTLP KISKEEFGLI FDELDDTRDF KINKDEFADL CQAIALRFQK EEVPSLFEHF PQIY HSALS QQLRAFVRSP NFGYAISFIL IINFIAVVVE TTLNIEESSA QKPWQVAEFV FGWIYVLEMA LKIYTYGFEN YWREG ANRF DFLVTWVIVI GETATFITPD ENTFFSNGEW IRYLLLARML RLIRLLMNVQ RYRAFIATFI TLIPSLMPYL GTIFCV LCI YCSIGVQVFG GLVNAGNKKL FETELAEDDY LLFNFNDYPN GMVTLFNLLV MGNWQVWMES YKDLTGTWWS ITYFVSF YV ITILLLLNLV VAFVLEAFFT ELDLEEEEKC QGQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 50 mM Tris, 200 mM NaCl and 0.06% glycodiosgenin, 1 mM EDTA |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 68456 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: PROJECTION MATCHING |
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7tdd: |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X