+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SIVmac239.K180S SOSIP in complex with FZ019.2 Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Simian immunodeficiency virus / Simian immunodeficiency virus /  | |||||||||
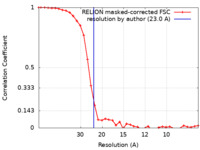
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Berndsen ZT | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Molecular insights into antibody-mediated protection against the prototypic simian immunodeficiency virus. Authors: Fangzhu Zhao / Zachary T Berndsen / Nuria Pedreño-Lopez / Alison Burns / Joel D Allen / Shawn Barman / Wen-Hsin Lee / Srirupa Chakraborty / Sandrasegaram Gnanakaran / Leigh M Sewall / ...Authors: Fangzhu Zhao / Zachary T Berndsen / Nuria Pedreño-Lopez / Alison Burns / Joel D Allen / Shawn Barman / Wen-Hsin Lee / Srirupa Chakraborty / Sandrasegaram Gnanakaran / Leigh M Sewall / Gabriel Ozorowski / Oliver Limbo / Ge Song / Peter Yong / Sean Callaghan / Jessica Coppola / Kim L Weisgrau / Jeffrey D Lifson / Rebecca Nedellec / Thomas B Voigt / Fernanda Laurino / Johan Louw / Brandon C Rosen / Michael Ricciardi / Max Crispin / Ronald C Desrosiers / Eva G Rakasz / David I Watkins / Raiees Andrabi / Andrew B Ward / Dennis R Burton / Devin Sok /   Abstract: SIVmac239 infection of macaques is a favored model of human HIV infection. However, the SIVmac239 envelope (Env) trimer structure, glycan occupancy, and the targets and ability of neutralizing ...SIVmac239 infection of macaques is a favored model of human HIV infection. However, the SIVmac239 envelope (Env) trimer structure, glycan occupancy, and the targets and ability of neutralizing antibodies (nAbs) to protect against SIVmac239 remain unknown. Here, we report the isolation of SIVmac239 nAbs that recognize a glycan hole and the V1/V4 loop. A high-resolution structure of a SIVmac239 Env trimer-nAb complex shows many similarities to HIV and SIVcpz Envs, but with distinct V4 features and an extended V1 loop. Moreover, SIVmac239 Env has a higher glycan shield density than HIV Env that may contribute to poor or delayed nAb responses in SIVmac239-infected macaques. Passive transfer of a nAb protects macaques from repeated intravenous SIVmac239 challenge at serum titers comparable to those described for protection of humans against HIV infection. Our results provide structural insights for vaccine design and shed light on antibody-mediated protection in the SIV model. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25623.map.gz emd_25623.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25623-v30.xml emd-25623-v30.xml emd-25623.xml emd-25623.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25623_fsc.xml emd_25623_fsc.xml | 3.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_25623.png emd_25623.png | 53.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25623 http://ftp.pdbj.org/pub/emdb/structures/EMD-25623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25623 | HTTPS FTP |
-Validation report
| Summary document |  emd_25623_validation.pdf.gz emd_25623_validation.pdf.gz | 367.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25623_full_validation.pdf.gz emd_25623_full_validation.pdf.gz | 366.8 KB | Display | |
| Data in XML |  emd_25623_validation.xml.gz emd_25623_validation.xml.gz | 7 KB | Display | |
| Data in CIF |  emd_25623_validation.cif.gz emd_25623_validation.cif.gz | 8.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25623 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25623 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25623 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25623 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25623.map.gz / Format: CCP4 / Size: 2.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25623.map.gz / Format: CCP4 / Size: 2.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.0195 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : SIVmac239.K180S SOSIP trimer in complex with 1 copy of FZ019.2 Fab
| Entire | Name: SIVmac239.K180S SOSIP trimer in complex with 1 copy of FZ019.2 Fab |
|---|---|
| Components |
|
-Supramolecule #1: SIVmac239.K180S SOSIP trimer in complex with 1 copy of FZ019.2 Fab
| Supramolecule | Name: SIVmac239.K180S SOSIP trimer in complex with 1 copy of FZ019.2 Fab type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Simian immunodeficiency virus Simian immunodeficiency virus |
-Macromolecule #1: SIVmac239.K180S SOSIP
| Macromolecule | Name: SIVmac239.K180S SOSIP / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  Simian immunodeficiency virus Simian immunodeficiency virus |
| Sequence | String: MGCLGNQLLI AILLLSVYGI YCTLYVTVFY GVPAWRNATI PLFCATKNRD TWGTTQCLPD NGDYSEMALN VTESFDAWNN TVTEQAIEDV WQLFETSIKP CVKLSPLCIT MRCNKSETDR WGLTKSITTT ASTTSTTASA KVDMVNETSS CIAQDNCTGL EQEQMISCKF ...String: MGCLGNQLLI AILLLSVYGI YCTLYVTVFY GVPAWRNATI PLFCATKNRD TWGTTQCLPD NGDYSEMALN VTESFDAWNN TVTEQAIEDV WQLFETSIKP CVKLSPLCIT MRCNKSETDR WGLTKSITTT ASTTSTTASA KVDMVNETSS CIAQDNCTGL EQEQMISCKF NMTGLKRDKS KEYNETWYSA DLVCEQGNNT GNESRCYMNH CNTSVIQESC DKHYWDAIRF RYCAPPGYAL LRCNDTNYSG FMPKCSKVVV SSCTRMMETQ TSTWFGFNGT RAENRTYIYW HGRDNRTIIS LNKYYNLTMK CRRPGNKTVL PVTIMSGLVF HSQPINDRPK QAWCWFGGKW KDAIKEVKQT IVKHPRYTGT NNTDKINLTA PGGGDPEVTF MWTNCRGEFL YCKMNWFLNW VEDRNTANQK PKEQHKRNYV PCHIRQIINT WHKVGKNVYL PPREGDLTCN STVTSLIANI DWIDGNQTNI TMSAEVAELY RLELGDYKLV EITPIGLAPT SCKRYTTGGT SRRRRRRGVF VLGFLGFLAT AGSAMGAASL TLTAQSRTLL AGIVQQQQQL LDVPKRQQEL LRLTVWGTKN LQTRVTAIEK YLKDQAQLNA WGCAFRQVCC TTVPWPNASL IPKWNNETWQ EWERKVDFLE ENITALLEEA QIQQEKNMYE LQKLNGSGHH HHHHHH |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #2: FZ019.2 Fab Heavy Chain
| Macromolecule | Name: FZ019.2 Fab Heavy Chain / type: other / ID: 2 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: EVQLVESGPG LVKPSETLSL TCTVSGGSVT ANYYWNWIRQ PPGKGLEWIG NVYGNALVNY YNPSLKSRIT ISKDIFKDQF FLKLSSVTAA DTAVYYCARR GGNQYNYFDV WGPGVLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV SWNSGALTSG ...String: EVQLVESGPG LVKPSETLSL TCTVSGGSVT ANYYWNWIRQ PPGKGLEWIG NVYGNALVNY YNPSLKSRIT ISKDIFKDQF FLKLSSVTAA DTAVYYCARR GGNQYNYFDV WGPGVLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV SWNSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKKVE PKSC |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #3: FZ019.2 Fab Light Chain
| Macromolecule | Name: FZ019.2 Fab Light Chain / type: other / ID: 3 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: QAGLTQPASL SASPGASASL TCTFSGGINV TGYHIFWYQQ KPGSPPRFLL RYKSDSDKGQ GSGVPSRFSG SKHVSANTGI FRISGLQSED EADYYCAIDH SSGDVFGSGT KLTVLGQPKA APSVTLFPPS SEELQANKAT LVCLISDFYP GAVTVAWKAD SSPVKAGVET ...String: QAGLTQPASL SASPGASASL TCTFSGGINV TGYHIFWYQQ KPGSPPRFLL RYKSDSDKGQ GSGVPSRFSG SKHVSANTGI FRISGLQSED EADYYCAIDH SSGDVFGSGT KLTVLGQPKA APSVTLFPPS SEELQANKAT LVCLISDFYP GAVTVAWKAD SSPVKAGVET TTPSKQSNNK YAASSYLSLT PEQWKSHKSY SCQVTHEGST VEKTVAPTEC S |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Staining | Type: NEGATIVE / Material: uranyl formate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Detector mode: COUNTING / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)