[English] 日本語
 Yorodumi
Yorodumi- EMDB-25478: G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | |||||||||
 Map data Map data | G32Q4 Fab in complex with RBD | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | antibody / virus / immunity / VIRAL PROTEIN / VIRAL PROTEIN-Immune System complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell ...Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / membrane fusion / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / receptor ligand activity / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
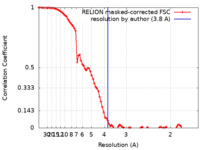
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Windsor IW / Tong P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Immunol / Year: 2022 Journal: Sci Immunol / Year: 2022Title: Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1. Authors: Ian W Windsor / Pei Tong / Olivia Lavidor / Ali Sanjari Moghaddam / Lindsay G A McKay / Avneesh Gautam / Yuezhou Chen / Elizabeth A MacDonald / Duck Kyun Yoo / Anthony Griffths / Duane R ...Authors: Ian W Windsor / Pei Tong / Olivia Lavidor / Ali Sanjari Moghaddam / Lindsay G A McKay / Avneesh Gautam / Yuezhou Chen / Elizabeth A MacDonald / Duck Kyun Yoo / Anthony Griffths / Duane R Wesemann / Stephen C Harrison /  Abstract: Neutralizing antibodies that recognize the SARS-CoV-2 spike glycoprotein are the principal host defense against viral invasion. Variants of SARS-CoV-2 bear mutations that allow escape from ...Neutralizing antibodies that recognize the SARS-CoV-2 spike glycoprotein are the principal host defense against viral invasion. Variants of SARS-CoV-2 bear mutations that allow escape from neutralization by many human antibodies, especially those in widely distributed ("public") classes. Identifying antibodies that neutralize these variants of concern and determining their prevalence are important goals for understanding immune protection. To determine the Delta and Omicron BA.1 variant specificity of B cell repertoires established by an initial Wuhan strain infection, we measured neutralization potencies of 73 antibodies from an unbiased survey of the early memory B cell response. Antibodies recognizing each of three previously defined epitopic regions on the spike receptor binding domain (RBD) varied in neutralization potency and variant-escape resistance. The ACE2 binding surface ("RBD-2") harbored the binding sites of neutralizing antibodies with the highest potency but with the greatest sensitivity to viral escape; two other epitopic regions on the RBD ("RBD-1" and "RBD-3") bound antibodies of more modest potency but greater breadth. The structures of several Fab:spike complexes that neutralized all five variants of concern tested, including one Fab each from the RBD-1, -2, and -3 clusters, illustrated the determinants of broad neutralization and showed that B cell repertoires can have specificities that avoid immune escape driven by public antibodies. The structure of the RBD-2 binding, broad neutralizer shows why it retains neutralizing activity for Omicron BA.1, unlike most others in the same public class. Our results correlate with real-world data on vaccine efficacy, which indicate mitigation of disease caused by Omicron BA.1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25478.map.gz emd_25478.map.gz | 48.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25478-v30.xml emd-25478-v30.xml emd-25478.xml emd-25478.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25478_fsc.xml emd_25478_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_25478.png emd_25478.png | 64.8 KB | ||
| Filedesc metadata |  emd-25478.cif.gz emd-25478.cif.gz | 5.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25478 http://ftp.pdbj.org/pub/emdb/structures/EMD-25478 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25478 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25478 | HTTPS FTP |
-Validation report
| Summary document |  emd_25478_validation.pdf.gz emd_25478_validation.pdf.gz | 553.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25478_full_validation.pdf.gz emd_25478_full_validation.pdf.gz | 553.1 KB | Display | |
| Data in XML |  emd_25478_validation.xml.gz emd_25478_validation.xml.gz | 10.3 KB | Display | |
| Data in CIF |  emd_25478_validation.cif.gz emd_25478_validation.cif.gz | 13.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25478 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25478 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25478 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25478 | HTTPS FTP |
-Related structure data
| Related structure data |  7swpMC  7swnC  7swoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25478.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25478.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | G32Q4 Fab in complex with RBD | ||||||||||||||||||||||||||||||||||||
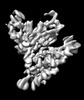
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : C98C7 Fab in complex with WT SARS-CoV-2 RBD
| Entire | Name: C98C7 Fab in complex with WT SARS-CoV-2 RBD |
|---|---|
| Components |
|
-Supramolecule #1: C98C7 Fab in complex with WT SARS-CoV-2 RBD
| Supramolecule | Name: C98C7 Fab in complex with WT SARS-CoV-2 RBD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.90157 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NLCPFGEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAS FSTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEVRQIAPG QTGKIADYN YKLPDDFTGC VIAWNSNNLD SKVGGNYNYL YRLFRKSNLK PFERDISTEI YQAGSTPCNG VEGFNCYFPL Q SYGFQPTN ...String: NLCPFGEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAS FSTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEVRQIAPG QTGKIADYN YKLPDDFTGC VIAWNSNNLD SKVGGNYNYL YRLFRKSNLK PFERDISTEI YQAGSTPCNG VEGFNCYFPL Q SYGFQPTN GVGYQPYRVV VLSFELLHAP ATVCGPK UniProtKB: Spike glycoprotein |
-Macromolecule #2: G32Q4 Fab heavy chain
| Macromolecule | Name: G32Q4 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.261164 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVESGGG VVQPGRSLRL SCAASGFTFS RYAMQWVRQA PGKGLEWVAV ISYDGNNRYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCARG PPGYYDSSGY YQTPEYFQHW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK D YFPEPVTV ...String: QVQLVESGGG VVQPGRSLRL SCAASGFTFS RYAMQWVRQA PGKGLEWVAV ISYDGNNRYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCARG PPGYYDSSGY YQTPEYFQHW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK D YFPEPVTV SWNSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKRVE PKSCDK |
-Macromolecule #3: G32Q4 Fab light chain
| Macromolecule | Name: G32Q4 Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.804156 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSV SGAPGQRVTI SCTGSSSNIG VGYDVHWYQQ FPGTVPKLLI YGNSNRPSGV PDRFSGSKSG TSASLAITGL QAEDEADYY CQSYDSSLSG VVFGGGTKLT VLGQPKAAPS VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADSSP V KAGVETTT ...String: QSVLTQPPSV SGAPGQRVTI SCTGSSSNIG VGYDVHWYQQ FPGTVPKLLI YGNSNRPSGV PDRFSGSKSG TSASLAITGL QAEDEADYY CQSYDSSLSG VVFGGGTKLT VLGQPKAAPS VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADSSP V KAGVETTT PSKQSNNKYA ASSYLSLTPE QWKSHRSYSC QVTHEGSTVE KTVAPTECS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 52.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)