+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Chitin Synthase 2 from Candida albicans at the apo state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | chitin synthesis / glycosyltransferase / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information: / chitin synthase / chitin synthase activity / cell septum / cell periphery / membrane Similarity search - Function | |||||||||
| Biological species |  Candida albicans (yeast) Candida albicans (yeast) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.95 Å | |||||||||
 Authors Authors | Ren Z / Chhetri A | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structural basis for inhibition and regulation of a chitin synthase from Candida albicans. Authors: Zhenning Ren / Abhishek Chhetri / Ziqiang Guan / Yang Suo / Kenichi Yokoyama / Seok-Yong Lee /  Abstract: Chitin is an essential component of the fungal cell wall. Chitin synthases (Chss) catalyze chitin formation and translocation across the membrane and are targets of antifungal agents, including ...Chitin is an essential component of the fungal cell wall. Chitin synthases (Chss) catalyze chitin formation and translocation across the membrane and are targets of antifungal agents, including nikkomycin Z and polyoxin D. Lack of structural insights into the action of these inhibitors on Chs has hampered their further development to the clinic. We present the cryo-EM structures of Chs2 from Candida albicans (CaChs2) in the apo, substrate-bound, nikkomycin Z-bound, and polyoxin D-bound states. CaChs2 adopts a unique domain-swapped dimer configuration where a conserved motif in the domain-swapped region controls enzyme activity. CaChs2 has a dual regulation mechanism where the chitin translocation tunnel is closed by the extracellular gate and plugged by a lipid molecule in the apo state to prevent non-specific leak. Analyses of substrate and inhibitor binding provide insights into the chemical logic of Chs inhibition, which can guide Chs-targeted antifungal development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25432.map.gz emd_25432.map.gz | 4.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25432-v30.xml emd-25432-v30.xml emd-25432.xml emd-25432.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25432.png emd_25432.png | 14.7 KB | ||
| Filedesc metadata |  emd-25432.cif.gz emd-25432.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25432 http://ftp.pdbj.org/pub/emdb/structures/EMD-25432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25432 | HTTPS FTP |
-Validation report
| Summary document |  emd_25432_validation.pdf.gz emd_25432_validation.pdf.gz | 408 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25432_full_validation.pdf.gz emd_25432_full_validation.pdf.gz | 407.6 KB | Display | |
| Data in XML |  emd_25432_validation.xml.gz emd_25432_validation.xml.gz | 5.9 KB | Display | |
| Data in CIF |  emd_25432_validation.cif.gz emd_25432_validation.cif.gz | 6.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25432 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25432 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25432 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25432 | HTTPS FTP |
-Related structure data
| Related structure data |  7stlMC  7stmC  7stnC  7stoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25432.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25432.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
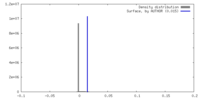
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Chitin synthase 2 from Candida albicans in the apo state
| Entire | Name: Chitin synthase 2 from Candida albicans in the apo state |
|---|---|
| Components |
|
-Supramolecule #1: Chitin synthase 2 from Candida albicans in the apo state
| Supramolecule | Name: Chitin synthase 2 from Candida albicans in the apo state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Candida albicans (yeast) Candida albicans (yeast) |
-Macromolecule #1: Chitin synthase
| Macromolecule | Name: Chitin synthase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: chitin synthase |
|---|---|
| Source (natural) | Organism:  Candida albicans (yeast) Candida albicans (yeast) |
| Molecular weight | Theoretical: 119.298227 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSYNNPNNSN SHLRPHAYNN SRRDDSDGDE SSIEFLNQRS NTPLTQGTYN YHNTSTNSLN FQQPEPIYRN QTRTSLSDSY YDHPIFDTS QTQIQPPHDN PFTESYEMTD TSYQGNDHHY RTGQPNHLMN PTYNQAFIPH VYDEEDNDEQ EYDQRIQYNQ F QGDHFDLA ...String: MSYNNPNNSN SHLRPHAYNN SRRDDSDGDE SSIEFLNQRS NTPLTQGTYN YHNTSTNSLN FQQPEPIYRN QTRTSLSDSY YDHPIFDTS QTQIQPPHDN PFTESYEMTD TSYQGNDHHY RTGQPNHLMN PTYNQAFIPH VYDEEDNDEQ EYDQRIQYNQ F QGDHFDLA AISYADDESQ SQLDYVPTER VIPEGEEEEE EGETSFEKEP GSETISGPFG EERSFEEPPP QQEVRSKKLT RA TGLNGHL VLDCPVADEL LSKFPDYNPA EKSGGLSREF AFMRYTAVTC GPSNFYRDAY ILRPVHYPIP RQTELMIVIT MYN EDDILL GRTLKGVFKN IKYLESKARS STWGKDSWKK IVVCIVSDGR TKINERAQAL LAGLGVYQEG LAKSRVDDKK VQAH MFEYT TRVGISKVTD DVVKLTTEKV VPVQMLFCLK ETNAKKINSH RWCFQAIGQV LDPKIVVLLD CGTQPSGRSL YELWK EFDR DHRVAGACGE ITTSLKKRQM ITNPLVYGQN FEYKISNILD KPTESSFGFI SVLPGAFSAY RFIALQNDIN GVGPLE KYF KGEFLHSSGE LDPNDDEFQM KHLMLKEEAG IFTSNMYLAE DRILCFELVA KRGCNWLLRY CKSARAETDV PEGLAEF IL QRRRWLNGSF FAAIYSLVHF YKVWTSSHSF GRKIFLHIEF FYQLINLIVS WFSIGSYFLV FRILTTSLGD KALGFAPG K ILSVIFLWLY LASIVTTFVL SFGNKPKGTE KFYVTIVIFF AILMAYMIFA AIFMAVHSIQ DIYRSGTRIT VSLFFQNSE FRDLVVATSS TYALYFLASF LYFEPWHMFT SFVQYILLSP SYVNVLNIYA FCNIDDISWG TKGEVGGKSL GEAKLREDGT FDVSVPISK EQINQSYLDQ LEKIRDPAPP EEKVLVTNTE DYYAFIRSMT VLVWMFTNFV VIALVLETGG FNQFVEATDL A NLKSNRAA VFLTVILWTV AFMALFRFIG CIYYLITRLG REIKASEHAT KANSLEVLFQ GPDYKDDDDK AHHHHHHHHH H UniProtKB: chitin synthase |
-Macromolecule #2: 1,2-Distearoyl-sn-glycerophosphoethanolamine
| Macromolecule | Name: 1,2-Distearoyl-sn-glycerophosphoethanolamine / type: ligand / ID: 2 / Number of copies: 8 / Formula: 3PE |
|---|---|
| Molecular weight | Theoretical: 748.065 Da |
| Chemical component information |  ChemComp-3PE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: From Ab initio |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.95 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 385452 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)