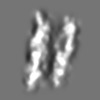[English] 日本語
 Yorodumi
Yorodumi- EMDB-25355: Tertiary structure of an individual particle of self-folding RNA ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tertiary structure of an individual particle of self-folding RNA polymer (particle #083) | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Single molecule structure / Individual Particle cryo-Electron Tomography / IPET / Cryo-ET / tertiary structure / Structural Flexibility / Self-folding mechanism / RNA origami / RNA | ||||||||||||||||||
| Biological species | synthetic construct (others) | ||||||||||||||||||
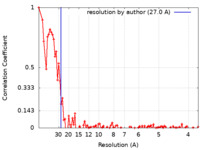
| Method | electron tomography / cryo EM / Resolution: 27.0 Å | ||||||||||||||||||
 Authors Authors | Liu J / Ren G | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation | Journal: bioRxiv / Year: 2023 Title: Tertiary structure of single-instant RNA molecule reveals folding landscape. Authors: Jianfang Liu / Ewan K S McRae / Meng Zhang / Cody Geary / Ebbe Sloth Andersen / Gang Ren Abstract: The folding of RNA and protein molecules during their synthesis is a crucial self-assembly process that nature employs to convert genetic information into the complex molecular machinery that ...The folding of RNA and protein molecules during their synthesis is a crucial self-assembly process that nature employs to convert genetic information into the complex molecular machinery that supports life. Misfolding events are the cause of several diseases, and the folding pathway of central biomolecules, such as the ribosome, is strictly regulated by programmed maturation processes and folding chaperones. However, the dynamic folding processes are challenging to study because current structure determination methods heavily rely on averaging, and existing computational methods do not efficiently simulate non-equilibrium dynamics. Here we utilize individual-particle cryo-electron tomography (IPET) to investigate the folding landscape of a rationally designed RNA origami 6-helix bundle that undergoes slow maturation from a "young" to "mature" conformation. By optimizing the IPET imaging and electron dose conditions, we obtain 3D reconstructions of 120 individual particles at resolutions ranging from 23-35 Å, enabling us first-time to observe individual RNA helices and tertiary structures without averaging. Statistical analysis of 120 tertiary structures confirms the two main conformations and suggests a possible folding pathway driven by helix-helix compaction. Studies of the full conformational landscape reveal both trapped states, misfolded states, intermediate states, and fully compacted states. The study provides novel insight into RNA folding pathways and paves the way for future studies of the energy landscape of molecular machines and self-assembly processes. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25355.map.gz emd_25355.map.gz | 6.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25355-v30.xml emd-25355-v30.xml emd-25355.xml emd-25355.xml | 13.3 KB 13.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25355_fsc.xml emd_25355_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_25355.png emd_25355.png | 78.8 KB | ||
| Filedesc metadata |  emd-25355.cif.gz emd-25355.cif.gz | 5.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25355 http://ftp.pdbj.org/pub/emdb/structures/EMD-25355 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25355 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25355 | HTTPS FTP |
-Validation report
| Summary document |  emd_25355_validation.pdf.gz emd_25355_validation.pdf.gz | 305.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25355_full_validation.pdf.gz emd_25355_full_validation.pdf.gz | 304.9 KB | Display | |
| Data in XML |  emd_25355_validation.xml.gz emd_25355_validation.xml.gz | 10.2 KB | Display | |
| Data in CIF |  emd_25355_validation.cif.gz emd_25355_validation.cif.gz | 13.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25355 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25355 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25355 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25355 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25355.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25355.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.88 Å | ||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : RNA Origami 6 Helix Bundle with Clasp (6HBC)
| Entire | Name: RNA Origami 6 Helix Bundle with Clasp (6HBC) |
|---|---|
| Components |
|
-Supramolecule #1: RNA Origami 6 Helix Bundle with Clasp (6HBC)
| Supramolecule | Name: RNA Origami 6 Helix Bundle with Clasp (6HBC) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The RNA was transcribed from linearized DNA template using T7 RNA polymerase purified in house. |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 230 KDa |
-Macromolecule #1: RNA Origami 6 Helix Bundle with Clasp (6HBC)
| Macromolecule | Name: RNA Origami 6 Helix Bundle with Clasp (6HBC) / type: rna / ID: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: GGGAGAGUAC UAUUCAGAUG CAGACCGCAA GUUCAGAGCG GUUUGCAUCU AGGGUACGUU UUCGAACGUA UCCUCCGACU AAGUGUAUUC GUAUACUUAG UGCCUUGUGC CUGCUUCGGC AGGCAUGACC CAAAUGUGCC UUUCGGGGCA CAUUUCCGGU CAUCCAAGUU ...String: GGGAGAGUAC UAUUCAGAUG CAGACCGCAA GUUCAGAGCG GUUUGCAUCU AGGGUACGUU UUCGAACGUA UCCUCCGACU AAGUGUAUUC GUAUACUUAG UGCCUUGUGC CUGCUUCGGC AGGCAUGACC CAAAUGUGCC UUUCGGGGCA CAUUUCCGGU CAUCCAAGUU CGCUUGGGUG AUGCGGGCGU AUAGGUUCGU CUAUACGUCC GCGUUUUCCG AGAAGAGGUA ACUCGGGAAA CCGGUCCACG UGACAAAGGU AGAGUUACGU GGAGGGAGCA GCUGCAAAGG GAUAAUGCAG UUGCUGGCUG GAUGCCAGAA CUCACGACUG GCAUCUACGG GGAUGGUGCU CUCCCAAUUC UCCAUUUACC GCCGAAUCGA CCCCAACGUG AGAGGGGUCG GUUCCCCGAG CAUAGACCAA UAUCCCAGGU UUAUGCUCCC CAACGCUGGA CGAACUACCU ACGUCUAGCG UUCCGGCAAA UGAGUCAAUA CCUCAGACUU AUUUGCGGUG CCUGAGCCUA AACUGAACAU GGGUUCAGGC AUCUUGGCUC CAGUUCGCUG GAGCCGACGG UAGCGCUGCG UUCGCGCAGU GCUAGGGAGC AUCCGUUUUC GAGCGGAUGC UGGGCGGUUG CCUGUUCGCA GGCAAUCGGG CCUACUCAUG AUUCGUCAUG AGUGGUGACA GCGUGAUGUU CGCAUUACGC UGUCGGGUAG AUGGAGAAUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: LEICA EM GP | ||||||||||||
| Details | In vitro transcribed RNA was purified by size exclusion chromatography and spin concentrated in amicon spin columns. | ||||||||||||
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 21 / Average exposure time: 0.88 sec. / Average electron dose: 8.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












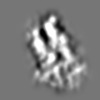




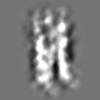




















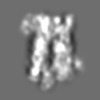
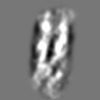




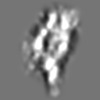
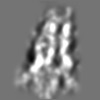





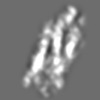






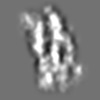


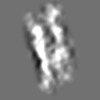
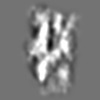


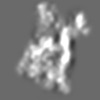

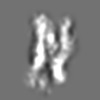

















































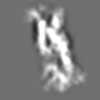


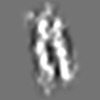


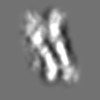

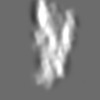



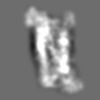
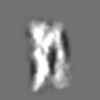





















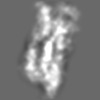


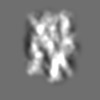





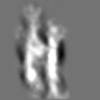



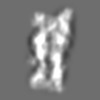




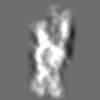

 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)