+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of type I-D Cascade bound to a dsDNA target | |||||||||
 Map data Map data | Sharpened Map of CRISPR-Cas type I-D complex bound to a dsDNA target. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / Complex / Ribonucleoprotein complex / type I-D / type ID / type I / cyanobacteria / synechocystis / RNA binding protein / DNA binding protein / DNA BINDING PROTEIN-DNA-RNA complex | |||||||||
| Function / homology | CRISPR-associated protein Csc1 / CRISPR associated protein Csc3 / CRISPR-associated protein Csc2 / Csc2 Crispr / Slr7013 protein / Slr7012 protein / CRISPR-associated protein Csc3 Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
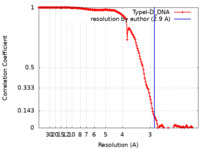
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Schwartz EA / Taylor DW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural rearrangements allow nucleic acid discrimination by type I-D Cascade. Authors: Evan A Schwartz / Tess M McBride / Jack P K Bravo / Daniel Wrapp / Peter C Fineran / Robert D Fagerlund / David W Taylor /   Abstract: CRISPR-Cas systems are adaptive immune systems that protect prokaryotes from foreign nucleic acids, such as bacteriophages. Two of the most prevalent CRISPR-Cas systems include type I and type III. ...CRISPR-Cas systems are adaptive immune systems that protect prokaryotes from foreign nucleic acids, such as bacteriophages. Two of the most prevalent CRISPR-Cas systems include type I and type III. Interestingly, the type I-D interference proteins contain characteristic features of both type I and type III systems. Here, we present the structures of type I-D Cascade bound to both a double-stranded (ds)DNA and a single-stranded (ss)RNA target at 2.9 and 3.1 Å, respectively. We show that type I-D Cascade is capable of specifically binding ssRNA and reveal how PAM recognition of dsDNA targets initiates long-range structural rearrangements that likely primes Cas10d for Cas3' binding and subsequent non-target strand DNA cleavage. These structures allow us to model how binding of the anti-CRISPR protein AcrID1 likely blocks target dsDNA binding via competitive inhibition of the DNA substrate engagement with the Cas10d active site. This work elucidates the unique mechanisms used by type I-D Cascade for discrimination of single-stranded and double stranded targets. Thus, our data supports a model for the hybrid nature of this complex with features of type III and type I systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24974.map.gz emd_24974.map.gz | 290.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24974-v30.xml emd-24974-v30.xml emd-24974.xml emd-24974.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24974_fsc.xml emd_24974_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_24974.png emd_24974.png | 52.4 KB | ||
| Filedesc metadata |  emd-24974.cif.gz emd-24974.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24974 http://ftp.pdbj.org/pub/emdb/structures/EMD-24974 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24974 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24974 | HTTPS FTP |
-Validation report
| Summary document |  emd_24974_validation.pdf.gz emd_24974_validation.pdf.gz | 365.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24974_full_validation.pdf.gz emd_24974_full_validation.pdf.gz | 365.2 KB | Display | |
| Data in XML |  emd_24974_validation.xml.gz emd_24974_validation.xml.gz | 14.5 KB | Display | |
| Data in CIF |  emd_24974_validation.cif.gz emd_24974_validation.cif.gz | 19.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24974 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24974 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24974 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24974 | HTTPS FTP |
-Related structure data
| Related structure data |  7sbaMC  7sbbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24974.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24974.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened Map of CRISPR-Cas type I-D complex bound to a dsDNA target. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Type I-D Cascade bound to a dsDNA target
| Entire | Name: Type I-D Cascade bound to a dsDNA target |
|---|---|
| Components |
|
-Supramolecule #1: Type I-D Cascade bound to a dsDNA target
| Supramolecule | Name: Type I-D Cascade bound to a dsDNA target / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 480 KDa |
-Macromolecule #1: Cas7d
| Macromolecule | Name: Cas7d / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.527109 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLDSLKSQFQ PSFPRLASGH YVHFLMLRHS QSFPVFQTDG VLNTTRTQAG LLEKTDQLSR LVMFKRKQTT PERLAGRELL RNLGLTSAD KSAKNLCEYN GEGSCKQCPD CILYGFAIGD SGSERSKVYS DSAFSLGAYE QSHRSFTFNA PFEGGTMSEA G VMRSAINE ...String: MLDSLKSQFQ PSFPRLASGH YVHFLMLRHS QSFPVFQTDG VLNTTRTQAG LLEKTDQLSR LVMFKRKQTT PERLAGRELL RNLGLTSAD KSAKNLCEYN GEGSCKQCPD CILYGFAIGD SGSERSKVYS DSAFSLGAYE QSHRSFTFNA PFEGGTMSEA G VMRSAINE LDHILPEVTF PTVESLRDAT YEGFIYVLGN LLRTKRYGAQ ESRTGTMKNH LVGIVFADGE IFSNLHLTQA LY DQMGGEL NKPISELCET AATVAQDLLN KEPVRKSELI FGAHLDTLLQ EVNDIYQNDA ELTKLLGSLY QQTQDYATEF GAL SGGKKK AKS UniProtKB: Slr7012 protein |
-Macromolecule #2: Cas5d
| Macromolecule | Name: Cas5d / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.942748 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTKIYRCKLT LHDNVFFASR EMGILYETEK YFHNWALSYA FFKGTIIPHP YGLVGQNAQT PAYLDRDREQ NLLHLNDSGI YVFPAQPIH WSYQINTFKA AQSAYYGRSV QFGGKGATKN YPINYGRAKE LAVGSEFLTY IVSQKELDLP VWIRLGKWSS K IRVEVEAI ...String: MTKIYRCKLT LHDNVFFASR EMGILYETEK YFHNWALSYA FFKGTIIPHP YGLVGQNAQT PAYLDRDREQ NLLHLNDSGI YVFPAQPIH WSYQINTFKA AQSAYYGRSV QFGGKGATKN YPINYGRAKE LAVGSEFLTY IVSQKELDLP VWIRLGKWSS K IRVEVEAI APDQIKTASG VYVCNHPLNP LDCPANQQIL LYNRVVMPPS SLFSQSQLQG DYWQIDRNTF LPQGFHYGAT TA IAQDSPQ LSLLDTN UniProtKB: Slr7013 protein |
-Macromolecule #3: Cas10d
| Macromolecule | Name: Cas10d / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 112.067508 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTLLQTLLI RTLSEQKDYI LLEYFQTILP ALEEHFGNTS GLGGSFISHQ KHFGTQGYDT EKAKKMAQGF AKKGDQTLAA HILNALLTT WNVMQELEFP LNDIERRLLC LGITLHDYDK HCHAQDMAAP EPDNIQEIIN ICLELGKRLN FDEFWADWRD Y IAEISYLA ...String: MTTLLQTLLI RTLSEQKDYI LLEYFQTILP ALEEHFGNTS GLGGSFISHQ KHFGTQGYDT EKAKKMAQGF AKKGDQTLAA HILNALLTT WNVMQELEFP LNDIERRLLC LGITLHDYDK HCHAQDMAAP EPDNIQEIIN ICLELGKRLN FDEFWADWRD Y IAEISYLA QNTHGKQHTN LISSNWSNAG YPFTIKERKL DHPLRHLLTF GDVAVHLSSP HDLVSSTMGD RLRDLLNRLG IE KRFVYHH LRDTTGILSN AIHNVILRTV QKLDWKPLLF FAQGVIYFAP QDTEIPERNE IKQIVWQGIS QELGKKMSAG DVG FKRDGK GLKVSPQTSE LLAAADIVRI LPQVISVKVN NAKSPATPKR LEKLELGDAE REKLYEVADL RCDRLAELLG LVQK EIFLL PEPFIEWVLK DLELTSVIMP EETQVQSGGV NYGWYRVAAH YVANHATWDL EEFQEFLQGF GDRLATWAEE EGYFA EHQS PTRQIFEDYL DRYLEIQGWE SDHQAFIQEL ENYVNAKTKK SKQPICSLSS GEFPSEDQMD SVVLFKPQQY SNKNPL GGG QIKRGISKIW SLEMLLRQAF WSVPSGKFED QQPIFIYLYP AYVYAPQVVE AIRELVYGIA SVNLWDVRKH WVNNKMD LT SLKSLPWLNE EVEAGTNAQL KYTKEDLPFL ATVYTTTREK TDTDAWVKPA FLALLLPYLL GVKAIATRSM VPLYRSDQ D FRESIHLDGV AGFWSLLGIP TDLRVEDITP ALNKLLAIYT LHLAARSSPP KARWQDLPKT VQEVMTDVLN VFALAEQGL RREKRDRPYE SEVTEYWQFA ELFSQGNIVM TEKLKLTKRL VEEYRRFYQV ELSKKPSTHA ILLPLSKALE QILSVPDDWD EEELILQGS GQLQAALDRQ EVYTRPIIKD KSVAYETRQL QELEAIQIFM TTCVRDLFGE MCKGDRAILQ EQRNRIKSGA E FAYRLLAL EAQQNQN UniProtKB: CRISPR-associated protein Csc3 |
-Macromolecule #4: Cas11d
| Macromolecule | Name: Cas11d / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.024494 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTEKLKLTKR LVEEYRRFYQ VELSKKPSTH AILLPLSKAL EQILSVPDDW DEEELILQGS GQLQAALDRQ EVYTRPIIKD KSVAYETRQ LQELEAIQIF MTTCVRDLFG EMCKGDRAIL QEQRNRIKSG AEFAYRLLAL EAQQNQN UniProtKB: CRISPR-associated protein Csc3 |
-Macromolecule #5: DNA target strand
| Macromolecule | Name: DNA target strand / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.884222 KDa |
| Sequence | String: (DA)(DC)(DA)(DA)(DC)(DA)(DA)(DT)(DC)(DA) (DA)(DC)(DG)(DT)(DG)(DA) |
-Macromolecule #6: DNA non-target strand
| Macromolecule | Name: DNA non-target strand / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 3.947581 KDa |
| Sequence | String: (DC)(DA)(DC)(DG)(DT)(DT)(DG)(DA)(DT)(DT) (DT)(DT)(DT) |
-Macromolecule #7: crRNA
| Macromolecule | Name: crRNA / type: rna / ID: 7 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.699081 KDa |
| Sequence | String: ACUGAAACGA UUGUUGUGCC CCUGGCGGUC GCUUUCAAUG CCU GENBANK: GENBANK: CP073020.1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.125 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: C-flat / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Blot force 0, blot time 4.5s, no wait time between sample application and blotting.. | |||||||||||||||
| Details | Monodisperse sample of elongated particles. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average exposure time: 3.0 sec. / Average electron dose: 41.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Model was built de novo using coot then flexibly refined using ISOLDE. |
|---|---|
| Output model |  PDB-7sba: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)