[English] 日本語
 Yorodumi
Yorodumi- EMDB-24824: Nanosandwich composite of two RNA-DNA hybrid nanotriangles bound ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24824 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nanosandwich composite of two RNA-DNA hybrid nanotriangles bound at a streptavidin core | |||||||||
 Map data Map data | Nano-sandwich composite consisting of two RNA-DNA hybrid nanotriangles bound to a streptavidin core. | |||||||||
 Sample Sample |
| |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.8 Å | |||||||||
 Authors Authors | Hermann T / Chen S / Xing L / Zhang D / Monferrer A | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2021 Journal: Nucleic Acids Res / Year: 2021Title: Nano-sandwich composite by kinetic trapping assembly from protein and nucleic acid. Authors: Shi Chen / Li Xing / Douglas Zhang / Alba Monferrer / Thomas Hermann /  Abstract: Design and preparation of layered composite materials alternating between nucleic acids and proteins has been elusive due to limitations in occurrence and geometry of interaction sites in natural ...Design and preparation of layered composite materials alternating between nucleic acids and proteins has been elusive due to limitations in occurrence and geometry of interaction sites in natural biomolecules. We report the design and kinetically controlled stepwise synthesis of a nano-sandwich composite by programmed noncovalent association of protein, DNA and RNA modules. A homo-tetramer protein core was introduced to control the self-assembly and precise positioning of two RNA-DNA hybrid nanotriangles in a co-parallel sandwich arrangement. Kinetically favored self-assembly of the circularly closed nanostructures at the protein was driven by the intrinsic fast folding ability of RNA corner modules which were added to precursor complex of DNA bound to the protein. The 3D architecture of this first synthetic protein-RNA-DNA complex was confirmed by fluorescence labeling and cryo-electron microscopy studies. The synthesis strategy for the nano-sandwich composite provides a general blueprint for controlled noncovalent assembly of complex supramolecular architectures from protein, DNA and RNA components, which expand the design repertoire for bottom-up preparation of layered biomaterials. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24824.map.gz emd_24824.map.gz | 6.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24824-v30.xml emd-24824-v30.xml emd-24824.xml emd-24824.xml | 8.8 KB 8.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24824.png emd_24824.png | 53.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24824 http://ftp.pdbj.org/pub/emdb/structures/EMD-24824 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24824 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24824 | HTTPS FTP |
-Validation report
| Summary document |  emd_24824_validation.pdf.gz emd_24824_validation.pdf.gz | 288.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24824_full_validation.pdf.gz emd_24824_full_validation.pdf.gz | 288.3 KB | Display | |
| Data in XML |  emd_24824_validation.xml.gz emd_24824_validation.xml.gz | 5.3 KB | Display | |
| Data in CIF |  emd_24824_validation.cif.gz emd_24824_validation.cif.gz | 6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24824 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24824 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24824 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24824 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24824.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24824.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nano-sandwich composite consisting of two RNA-DNA hybrid nanotriangles bound to a streptavidin core. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
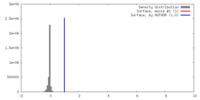
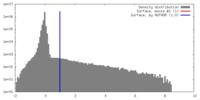
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Sandwich complex of two RNA-DNA hybrid nanotriangles bound at a s...
| Entire | Name: Sandwich complex of two RNA-DNA hybrid nanotriangles bound at a streptavidin core |
|---|---|
| Components |
|
-Supramolecule #1: Sandwich complex of two RNA-DNA hybrid nanotriangles bound at a s...
| Supramolecule | Name: Sandwich complex of two RNA-DNA hybrid nanotriangles bound at a streptavidin core type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Component - Concentration: 10.0 mM / Component - Name: HEPES |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: PROPANE / Chamber humidity: 95 % / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2100F |
|---|---|
| Image recording | Film or detector model: OTHER / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: GATAN 915 DOUBLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)