[English] 日本語
 Yorodumi
Yorodumi- EMDB-24636: Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees Celsius, in a closed state, class I | |||||||||
 Map data Map data | 4C_RTX_closed | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ligand-gating ion channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to capsazepine / negative regulation of establishment of blood-brain barrier / sensory perception of mechanical stimulus / excitatory extracellular ligand-gated monoatomic ion channel activity / peptide secretion / temperature-gated ion channel activity / cellular response to temperature stimulus / detection of chemical stimulus involved in sensory perception of pain / urinary bladder smooth muscle contraction / TRP channels ...response to capsazepine / negative regulation of establishment of blood-brain barrier / sensory perception of mechanical stimulus / excitatory extracellular ligand-gated monoatomic ion channel activity / peptide secretion / temperature-gated ion channel activity / cellular response to temperature stimulus / detection of chemical stimulus involved in sensory perception of pain / urinary bladder smooth muscle contraction / TRP channels / smooth muscle contraction involved in micturition / fever generation / thermoception / detection of temperature stimulus involved in thermoception / cellular response to acidic pH / response to pH / negative regulation of systemic arterial blood pressure / dendritic spine membrane / chloride channel regulator activity / glutamate secretion / monoatomic cation transmembrane transporter activity / negative regulation of heart rate / cellular response to ATP / response to pain / temperature homeostasis / cellular response to alkaloid / diet induced thermogenesis / cellular response to cytokine stimulus / ligand-gated monoatomic ion channel activity / behavioral response to pain / intracellularly gated calcium channel activity / detection of temperature stimulus involved in sensory perception of pain / negative regulation of mitochondrial membrane potential / calcium ion import across plasma membrane / monoatomic cation channel activity / extracellular ligand-gated monoatomic ion channel activity / sensory perception of pain / phosphatidylinositol binding / GABA-ergic synapse / phosphoprotein binding / microglial cell activation / cellular response to nerve growth factor stimulus / cellular response to growth factor stimulus / calcium ion transmembrane transport / calcium channel activity / lipid metabolic process / response to peptide hormone / calcium ion transport / positive regulation of nitric oxide biosynthetic process / transmembrane signaling receptor activity / sensory perception of taste / cellular response to tumor necrosis factor / cellular response to heat / response to heat / positive regulation of cytosolic calcium ion concentration / monoatomic ion transmembrane transport / protein homotetramerization / postsynaptic membrane / calmodulin binding / neuron projection / positive regulation of apoptotic process / external side of plasma membrane / neuronal cell body / dendrite / negative regulation of transcription by RNA polymerase II / ATP binding / identical protein binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | |||||||||
 Authors Authors | Kwon DH / Suo Y | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis. Authors: Do Hoon Kwon / Feng Zhang / Justin G Fedor / Yang Suo / Seok-Yong Lee /  Abstract: Single particle cryo-EM often yields multiple protein conformations within a single dataset, but experimentally deducing the temporal relationship of these conformers within a conformational ...Single particle cryo-EM often yields multiple protein conformations within a single dataset, but experimentally deducing the temporal relationship of these conformers within a conformational trajectory is not trivial. Here, we use thermal titration methods and cryo-EM in an attempt to obtain temporal resolution of the conformational trajectory of the vanilloid receptor TRPV1 with resiniferatoxin (RTx) bound. Based on our cryo-EM ensemble analysis, RTx binding to TRPV1 appears to induce intracellular gate opening first, followed by selectivity filter dilation, then pore loop rearrangement to reach the final open state. This apparent conformational wave likely arises from the concerted, stepwise, additive structural changes of TRPV1 over many subdomains. Greater understanding of the RTx-mediated long-range allostery of TRPV1 could help further the therapeutic potential of RTx, which is a promising drug candidate for pain relief associated with advanced cancer or knee arthritis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24636.map.gz emd_24636.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24636-v30.xml emd-24636-v30.xml emd-24636.xml emd-24636.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24636.png emd_24636.png | 65.7 KB | ||
| Filedesc metadata |  emd-24636.cif.gz emd-24636.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24636 http://ftp.pdbj.org/pub/emdb/structures/EMD-24636 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24636 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24636 | HTTPS FTP |
-Validation report
| Summary document |  emd_24636_validation.pdf.gz emd_24636_validation.pdf.gz | 526.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24636_full_validation.pdf.gz emd_24636_full_validation.pdf.gz | 526.4 KB | Display | |
| Data in XML |  emd_24636_validation.xml.gz emd_24636_validation.xml.gz | 6.2 KB | Display | |
| Data in CIF |  emd_24636_validation.cif.gz emd_24636_validation.cif.gz | 7.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24636 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24636 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24636 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24636 | HTTPS FTP |
-Related structure data
| Related structure data |  7rquMC  7rqvC  7rqwC  7rqxC  7rqyC  7rqzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24636.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24636.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 4C_RTX_closed | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Transient receptor potential cation channel subfamily V member 1
| Entire | Name: Transient receptor potential cation channel subfamily V member 1 |
|---|---|
| Components |
|
-Supramolecule #1: Transient receptor potential cation channel subfamily V member 1
| Supramolecule | Name: Transient receptor potential cation channel subfamily V member 1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 1
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 1 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 98.719758 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEQRASLDSE ESESPPQENS CLDPPDRDPN CKPPPVKPHI FTTRSRTRLF GKGDSEEASP LDCPYEEGGL ASCPIITVSS VLTIQRPGD GPASVRPSSQ DSVSAGEKPP RLYDRRSIFD AVAQSNCQEL ESLLPFLQRS KKRLTDSEFK DPETGKTCLL K AMLNLHNG ...String: MEQRASLDSE ESESPPQENS CLDPPDRDPN CKPPPVKPHI FTTRSRTRLF GKGDSEEASP LDCPYEEGGL ASCPIITVSS VLTIQRPGD GPASVRPSSQ DSVSAGEKPP RLYDRRSIFD AVAQSNCQEL ESLLPFLQRS KKRLTDSEFK DPETGKTCLL K AMLNLHNG QNDTIALLLD VARKTDSLKQ FVNASYTDSY YKGQTALHIA IERRNMTLVT LLVENGADVQ AAANGDFFKK TK GRPGFYF GELPLSLAAC TNQLAIVKFL LQNSWQPADI SARDSVGNTV LHALVEVADN TVDNTKFVTS MYNEILILGA KLH PTLKLE EITNRKGLTP LALAASSGKI GVLAYILQRE IHEPECRHLS RKFTEWAYGP VHSSLYDLSC IDTCEKNSVL EVIA YSSSE TPNRHDMLLV EPLNRLLQDK WDRFVKRIFY FNFFVYCLYM IIFTAAAYYR PVEGLPPYKL KNTVGDYFRV TGEIL SVSG GVYFFFRGIQ YFLQRRPSLK SLFVDSYSEI LFFVQSLFML VSVVLYFSQR KEYVASMVFS LAMGWTNMLY YTRGFQ QMG IYAVMIEKMI LRDLCRFMFV YLVFLFGFST AVVTLIEDGK NNSLPMESTP HKCRGSACKP GNSYNSLYST CLELFKF TI GMGDLEFTEN YDFKAVFIIL LLAYVILTYI LLLNMLIALM GETVNKIAQE SKNIWKLQRA ITILDTEKSF LKCMRKAF R SGKLLQVGFT PDGKDDYRWC FRVDEVNWTT WNTNVGIINE DPGNCEGVKR TLSFSLRSGR VSGRNWKNFA LVPLLRDAS TRDRHATQQE EVQLKHYTGS LKPEDAEVFK DSMVPGEKEN SLEVLFQGPD YKDDDDKAHH HHHHHHHH UniProtKB: Transient receptor potential cation channel subfamily V member 1 |
-Macromolecule #2: resiniferatoxin
| Macromolecule | Name: resiniferatoxin / type: ligand / ID: 2 / Number of copies: 4 / Formula: 6EU |
|---|---|
| Molecular weight | Theoretical: 628.708 Da |
| Chemical component information | 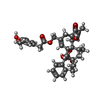 ChemComp-6EU: |
-Macromolecule #3: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
| Macromolecule | Name: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine / type: ligand / ID: 3 / Number of copies: 8 / Formula: LBN |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-LBN: |
-Macromolecule #4: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyl...
| Macromolecule | Name: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate type: ligand / ID: 4 / Number of copies: 20 / Formula: 6OU |
|---|---|
| Molecular weight | Theoretical: 717.996 Da |
| Chemical component information |  ChemComp-6OU: |
-Macromolecule #5: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol
| Macromolecule | Name: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol / type: ligand / ID: 5 / Number of copies: 4 / Formula: YFP |
|---|---|
| Molecular weight | Theoretical: 749.007 Da |
| Chemical component information | 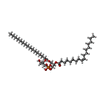 ChemComp-YFP: |
-Macromolecule #6: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 6 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 2427 / Average exposure time: 4.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 132 |
| Output model |  PDB-7rqu: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















