[English] 日本語
 Yorodumi
Yorodumi- EMDB-24389: Cryo-EM structure of the unliganded form of NLR family apoptosis ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the unliganded form of NLR family apoptosis inhibitory protein 5 (NAIP5) with partial LRR domain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Paidimuddala B / Cao J / Xie Q / Wu H / Zhang L | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Mechanism of NAIP-NLRC4 inflammasome activation revealed by cryo-EM structure of unliganded NAIP5. Authors: Bhaskar Paidimuddala / Jianhao Cao / Grady Nash / Qing Xie / Hao Wu / Liman Zhang /  Abstract: The nucleotide-binding domain (NBD), leucine rich repeat (LRR) domain containing protein family (NLR family) apoptosis inhibitory proteins (NAIPs) are cytosolic receptors that play critical roles in ...The nucleotide-binding domain (NBD), leucine rich repeat (LRR) domain containing protein family (NLR family) apoptosis inhibitory proteins (NAIPs) are cytosolic receptors that play critical roles in the host defense against bacterial infection. NAIPs interact with conserved bacterial ligands and activate the NLR family caspase recruitment domain containing protein 4 (NLRC4) to initiate the NAIP-NLRC4 inflammasome pathway. Here we found the process of NAIP activation is completely different from NLRC4. Our cryo-EM structure of unliganded mouse NAIP5 adopts an unprecedented wide-open conformation, with the nucleating surface fully exposed and accessible to recruit inactive NLRC4. Upon ligand binding, the winged helix domain (WHD) of NAIP5 undergoes roughly 20° rotation to form a steric clash with the inactive NLRC4, which triggers the conformational change of NLRC4 from inactive to active state. We also show the rotation of WHD places the 17-18 loop at a position that directly bind the active NLRC4 and stabilize the NAIP5-NLRC4 complex. Overall, these data provide structural mechanisms of inactive NAIP5, the process of NAIP5 activation and NAIP-dependent NLRC4 activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24389.map.gz emd_24389.map.gz | 57 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24389-v30.xml emd-24389-v30.xml emd-24389.xml emd-24389.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24389.png emd_24389.png | 64.1 KB | ||
| Others |  emd_24389_half_map_1.map.gz emd_24389_half_map_1.map.gz emd_24389_half_map_2.map.gz emd_24389_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24389 http://ftp.pdbj.org/pub/emdb/structures/EMD-24389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24389 | HTTPS FTP |
-Validation report
| Summary document |  emd_24389_validation.pdf.gz emd_24389_validation.pdf.gz | 656 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24389_full_validation.pdf.gz emd_24389_full_validation.pdf.gz | 655.5 KB | Display | |
| Data in XML |  emd_24389_validation.xml.gz emd_24389_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_24389_validation.cif.gz emd_24389_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24389 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24389 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24389 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24389 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24389.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24389.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0694 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_24389_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
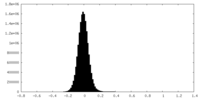
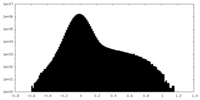
| Density Histograms |
-Half map: #1
| File | emd_24389_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
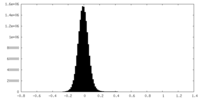
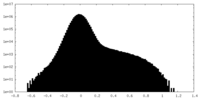
| Density Histograms |
- Sample components
Sample components
-Entire : Pre-liganded form of NAIP5
| Entire | Name: Pre-liganded form of NAIP5 |
|---|---|
| Components |
|
-Supramolecule #1: Pre-liganded form of NAIP5
| Supramolecule | Name: Pre-liganded form of NAIP5 / type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: NLR family) apoptosis inhibitory protein 5 (NAIP5)
| Macromolecule | Name: NLR family) apoptosis inhibitory protein 5 (NAIP5) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKL AEHGESSEDR ISEIDYEFLP ELSALLGVDA FQVAKSQEEE EHKERMKMKK GFNSQMRSEA KRLKTFETYD TFRSWTPQEM AAAGFYHTGV RLGVQCFCCS LILFGNSLRK LPIERHKKLR PECEFLQGKD VGNIGKYDIR VKRPEKMLRG GKARYHEEEA ...String: MDYKDDDDKL AEHGESSEDR ISEIDYEFLP ELSALLGVDA FQVAKSQEEE EHKERMKMKK GFNSQMRSEA KRLKTFETYD TFRSWTPQEM AAAGFYHTGV RLGVQCFCCS LILFGNSLRK LPIERHKKLR PECEFLQGKD VGNIGKYDIR VKRPEKMLRG GKARYHEEEA RLESFEDWPF YAHGTSPRV LSAAGFVFTG KRDTVQCFSC GGSLGNWEEG DDPWKEHAKW FPKCEFLQSK KSSEEIAQYI QSYEGFVHV TGEHFVKSWV RRELPMVSAY CNDSVFANEE LRMDMFKDWP QESPVGVEAL V RAGFFYTG KKDIVRCFSC GGCLEKWAEG DDPMEDHIKF FPECVFLQTL KSSAEVIPTL QS QYALPEA TETTRESNHG DAAAVHSTVV DLGRSEAQWF QEARSLSEQL RDNYTKATFR HMN LPEVCS SLGTDHLLSC DVSIISKHIS QPVQEALTIP EVFSNLNSVM CVEGETGSGK TTFL KRIAF LWASGCCPLL YRFQLVFYLS LSSITPDQGL ANIICAQLLG AGGCISEVCL SSSIQ QLQH QVLFLLDDYS GLASLPQALH TLITKNYLSR TCLLIAVHTN RVRDIRLYLG TSLEIQ EFP FYNTVSVLRK FFSHDIICVE KLIIYFIDNK DLQGVYKTPL FVAAVCTDWI QNASAQD KF QDVTLFQSYM QYLSLKYKAT AEPLQATVSS CGQLALTGLF SSCFEFNSDD LAEAGVDE D EKLTTLLMSK FTAQRLRPVY RFLGPLFQEF LAAVRLTELL SSDRQEDQDL GLYYLRQID SPLKAINSFN IFLYYVSSHS SSKAAPTVVS HLLQLVDEKE SLENMSENED YMKLHPQTFL WFQFVRGLW LVSPESSSSF VSEHLLRLAL IFAYESNTVA ECSPFILQFL RGKTLALRVL N LQYFRDHP ESLLLLRSLK VSINGNKMSS YVDYSFKTYF ENLQPPAIDE EYTSAFEHIS EW RRNFAQD EEIIKNYENI RPRALPDISE GYWKLSPKPC KIPKLEVQVN NTDAADQALL QVL MEVFSA SQSIEFRLFN SSGFLESICP ALELSKASVT KCSMSRLELS RAEQELLLTL PALQ SLEVS ETNQLPEQLF HNLHKFLGLK ELCVRLDGKP NVLSVLPREF PNLLHMEKLS IQTST ESDL SKLVKFIQNF PNLHVFHLKC DFLSNCESLM AVLASCKKLR EIEFSGRCFE AMTFVN ILP NFVSLKILNL KDQQFPDKET SEKFAQALGS LRNLEELLVP TGDGIHQVAK LIVRQCL QL PCLRVLTFHD ILDDDSVIEI ARAATSGGFQ KLENLDISMN HKITEEGYRN FFQALDNL P NLQELNICRN IPGRIQVQAT TVKALGQCVS RLPSLIRLHM LSWLLDEEDM KVINDVKER HPQSKRLIIF WKLIVPFSPV ILE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Concentration | 0.30 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 159513 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)