[English] 日本語
 Yorodumi
Yorodumi- EMDB-2419: 3-dimensional structure of the toxin-delivery particle antifeedin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2419 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3-dimensional structure of the toxin-delivery particle antifeeding prophage of Serratia entomophila | |||||||||
 Map data Map data | Reconstruction of the full Afp particle | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | tube-baseplate complex / tailocin / type VI secretion system / helical sheath / phage tail-like | |||||||||
| Biological species |  Serratia entomophila (bacteria) Serratia entomophila (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Heymann JB / Bartho JD / Rybakova D / Venugopal HP / Winkler DC / Sen A / Hurst MRH / Mitra AK | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2013 Journal: J Biol Chem / Year: 2013Title: Three-dimensional structure of the toxin-delivery particle antifeeding prophage of Serratia entomophila. Authors: J Bernard Heymann / Joseph D Bartho / Daria Rybakova / Hari P Venugopal / Dennis C Winkler / Anindito Sen / Mark R H Hurst / Alok K Mitra /   Abstract: The Serratia entomophila antifeeding prophage (Afp) is a bullet-shaped toxin-delivery apparatus similar to the R-pyocins of Pseudomonas aeruginosa. Morphologically it resembles the sheathed tail of ...The Serratia entomophila antifeeding prophage (Afp) is a bullet-shaped toxin-delivery apparatus similar to the R-pyocins of Pseudomonas aeruginosa. Morphologically it resembles the sheathed tail of bacteriophages such as T4, including a baseplate at one end. It also shares features with the type VI secretion systems. Cryo-electron micrographs of tilted Afp specimens (up to 60 degrees) were analyzed to determine the correct cyclic symmetry to overcome the limitation imposed by exclusively side views in nominally untilted specimens. An asymmetric reconstruction shows clear 6-fold cyclic symmetry contrary to a previous conclusion of 4-fold symmetry based on analysis of only the preferred side views (Sen, A., Rybakova, D., Hurst, M. R., and Mitra, A. K. (2010) J. Bacteriol. 192, 4522-4525). Electron tomography of negatively stained Afp revealed right-handed helical striations in many of the particles, establishing the correct hand. Higher quality micrographs of untilted specimens were processed to produce a reconstruction at 2.0-nm resolution with imposed 6-fold symmetry. The helical parameters of the sheath were determined to be 8.14 nm for the subunit rise along and 40.5° for the rotation angle around the helix. The sheath is similar to that of the T4 phage tail but with a different arrangement of the subdomain of the polymerizing sheath protein(s). The central tube is similar to the diameter and axial width of the Hcp1 hexamer of P. aeruginosa type VI secretion system. The tube extends through the baseplate into a needle resembling the "puncture device" of the T4 tail. The tube contains density that may be the toxin and/or a length-determining protein. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2419.map.gz emd_2419.map.gz | 20.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2419-v30.xml emd-2419-v30.xml emd-2419.xml emd-2419.xml | 9.2 KB 9.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_2419_fsc.xml emd_2419_fsc.xml | 8.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_2419.png emd_2419.png | 386.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2419 http://ftp.pdbj.org/pub/emdb/structures/EMD-2419 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2419 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2419 | HTTPS FTP |
-Validation report
| Summary document |  emd_2419_validation.pdf.gz emd_2419_validation.pdf.gz | 219.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2419_full_validation.pdf.gz emd_2419_full_validation.pdf.gz | 218.4 KB | Display | |
| Data in XML |  emd_2419_validation.xml.gz emd_2419_validation.xml.gz | 9.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2419 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2419 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2419 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2419 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2419.map.gz / Format: CCP4 / Size: 21.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2419.map.gz / Format: CCP4 / Size: 21.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of the full Afp particle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
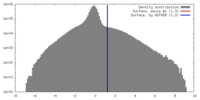
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Antifeeding prophage of Serratia entomophila
| Entire | Name: Antifeeding prophage of Serratia entomophila |
|---|---|
| Components |
|
-Supramolecule #1000: Antifeeding prophage of Serratia entomophila
| Supramolecule | Name: Antifeeding prophage of Serratia entomophila / type: sample / ID: 1000 / Details: Afp particle produced from 18 ORF products / Number unique components: 1 |
|---|
-Macromolecule #1: Antifeeding prophage
| Macromolecule | Name: Antifeeding prophage / type: protein_or_peptide / ID: 1 / Name.synonym: Afp Details: Product of the expression of 18 ORF's of the pADAP plasmid of Serratia entomophila Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Serratia entomophila (bacteria) Serratia entomophila (bacteria) |
| Recombinant expression | Organism:  Recombinant plasmid: pAF6-21, pASANFA1 |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 25 mM TBS, 130 mM NaCl |
|---|---|
| Staining | Type: NEGATIVE / Details: None (cryo) |
| Grid | Details: Quantifoil |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: HOMEMADE PLUNGER Method: Glow-discharge grid in amyl amine atmosphere Apply 5 ul sample, 4C, humidity 90% Blot Plunge |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Date | Feb 11, 2013 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 12.7 µm / Number real images: 113 / Bits/pixel: 16 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 41423 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.8 µm / Nominal defocus min: 1.6 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder: Gatan 626 / Specimen holder model: GATAN LIQUID NITROGEN |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera


 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)