[English] 日本語
 Yorodumi
Yorodumi- EMDB-23700: Full length alpha1 Glycine receptor in presence of 32uM Tetrahydr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full length alpha1 Glycine receptor in presence of 32uM Tetrahydrocannabinol | |||||||||
 Map data Map data | Primary Map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationtransmitter-gated monoatomic ion channel activity / Neurotransmitter receptors and postsynaptic signal transmission / extracellularly glycine-gated ion channel activity / extracellularly glycine-gated chloride channel activity / cellular response to ethanol / cellular response to zinc ion / regulation of neuron differentiation / neurotransmitter receptor activity / glycine binding / chloride channel complex ...transmitter-gated monoatomic ion channel activity / Neurotransmitter receptors and postsynaptic signal transmission / extracellularly glycine-gated ion channel activity / extracellularly glycine-gated chloride channel activity / cellular response to ethanol / cellular response to zinc ion / regulation of neuron differentiation / neurotransmitter receptor activity / glycine binding / chloride channel complex / ligand-gated monoatomic ion channel activity / transmembrane transporter complex / response to amino acid / monoatomic ion transport / chloride transmembrane transport / central nervous system development / cellular response to amino acid stimulus / transmembrane signaling receptor activity / perikaryon / postsynaptic membrane / neuron projection / dendrite / synapse / zinc ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
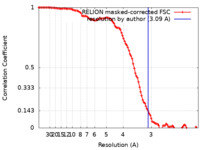
| Method | single particle reconstruction / cryo EM / Resolution: 3.09 Å | |||||||||
 Authors Authors | Kumar A / Chakrapani S | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs. Authors: Arvind Kumar / Sandip Basak / Shanlin Rao / Yvonne Gicheru / Megan L Mayer / Mark S P Sansom / Sudha Chakrapani /   Abstract: Glycinergic synapses play a central role in motor control and pain processing in the central nervous system. Glycine receptors (GlyRs) are key players in mediating fast inhibitory neurotransmission ...Glycinergic synapses play a central role in motor control and pain processing in the central nervous system. Glycine receptors (GlyRs) are key players in mediating fast inhibitory neurotransmission at these synapses. While previous high-resolution structures have provided insights into the molecular architecture of GlyR, several mechanistic questions pertaining to channel function are still unanswered. Here, we present Cryo-EM structures of the full-length GlyR protein complex reconstituted into lipid nanodiscs that are captured in the unliganded (closed), glycine-bound (open and desensitized), and allosteric modulator-bound conformations. A comparison of these states reveals global conformational changes underlying GlyR channel gating and modulation. The functional state assignments were validated by molecular dynamics simulations, and the observed permeation events are in agreement with the anion selectivity and conductance of GlyR. These studies provide the structural basis for gating, ion selectivity, and single-channel conductance properties of GlyR in a lipid environment. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23700.map.gz emd_23700.map.gz | 92.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23700-v30.xml emd-23700-v30.xml emd-23700.xml emd-23700.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23700_fsc.xml emd_23700_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_23700.png emd_23700.png | 55.9 KB | ||
| Others |  emd_23700_additional_1.map.gz emd_23700_additional_1.map.gz emd_23700_additional_2.map.gz emd_23700_additional_2.map.gz emd_23700_half_map_1.map.gz emd_23700_half_map_1.map.gz emd_23700_half_map_2.map.gz emd_23700_half_map_2.map.gz | 80.6 MB 96 MB 80.9 MB 80.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23700 http://ftp.pdbj.org/pub/emdb/structures/EMD-23700 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23700 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23700 | HTTPS FTP |
-Validation report
| Summary document |  emd_23700_validation.pdf.gz emd_23700_validation.pdf.gz | 551.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23700_full_validation.pdf.gz emd_23700_full_validation.pdf.gz | 551.3 KB | Display | |
| Data in XML |  emd_23700_validation.xml.gz emd_23700_validation.xml.gz | 17.9 KB | Display | |
| Data in CIF |  emd_23700_validation.cif.gz emd_23700_validation.cif.gz | 23.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23700 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23700 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23700 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23700 | HTTPS FTP |
-Related structure data
| Related structure data |  7m6mMC  7m6nC  7m6oC  7m6pC  7m6qC  7m6rC  7m6sC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23700.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23700.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: maps generated in the final rounds of 3D-autorefinement in relion 3.1
| File | emd_23700_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | maps generated in the final rounds of 3D-autorefinement in relion 3.1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: postprocessing maps generated ysing half maps and softmask...
| File | emd_23700_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocessing maps generated ysing half maps and softmask in relion 3.1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: maps reconstructed independently each using half of the...
| File | emd_23700_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | maps reconstructed independently each using half of the experimental data | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: maps reconstructed independently each using half of the...
| File | emd_23700_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | maps reconstructed independently each using half of the experimental data | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Glycine receptor subunit alpha Z1
| Entire | Name: Glycine receptor subunit alpha Z1 |
|---|---|
| Components |
|
-Supramolecule #1: Glycine receptor subunit alpha Z1
| Supramolecule | Name: Glycine receptor subunit alpha Z1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 250 kDa/nm |
-Macromolecule #1: Glycine receptor subunit alphaZ1
| Macromolecule | Name: Glycine receptor subunit alphaZ1 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.821711 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFALGIYLWE TIVFFSLAAS QQAAARKAAS PMPPSEFLDK LMGKVSGYDA RIRPNFKGPP VNVTCNIFIN SFGSIAETTM DYRVNIFLR QQWNDPRLAY SEYPDDSLDL DPSMLDSIWK PDLFFANEKG ANFHEVTTDN KLLRISKNGN VLYSIRITLV L ACPMDLKN ...String: MFALGIYLWE TIVFFSLAAS QQAAARKAAS PMPPSEFLDK LMGKVSGYDA RIRPNFKGPP VNVTCNIFIN SFGSIAETTM DYRVNIFLR QQWNDPRLAY SEYPDDSLDL DPSMLDSIWK PDLFFANEKG ANFHEVTTDN KLLRISKNGN VLYSIRITLV L ACPMDLKN FPMDVQTCIM QLESFGYTMN DLIFEWDEKG AVQVADGLTL PQFILKEEKD LRYCTKHYNT GKFTCIEARF HL ERQMGYY LIQMYIPSLL IVILSWVSFW INMDAAPARV GLGITTVLTM TTQSSGSRAS LPKVSYVKAI DIWMAVCLLF VFS ALLEYA AVNFIARQHK ELLRFQRRRR HLKEDEAGDG RFSFAAYGMG PACLQAKDGM AIKGNNNNAP TSTNPPEKTV EEMR KLFIS RAKRIDTVSR VAFPLVFLIF NIFYWITYKI IRSEDIHKQ |
-Macromolecule #3: (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benz...
| Macromolecule | Name: (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol type: ligand / ID: 3 / Number of copies: 5 / Formula: TCI |
|---|---|
| Molecular weight | Theoretical: 314.462 Da |
| Chemical component information | 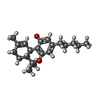 ChemComp-TCI: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK IV Details: 3.5 ul of 0.1 mg/ml protein solution was applied on a grid in the Vitrobot MkIV chamber set to 100% RH at 4 degC for 30s and then blotted for 2 s and plunged. | |||||||||
| Details | Full length Zebrafish GlyR alpha1 homopentamer reconsituted in Nanodisc |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 8 / Number real images: 9700 / Average exposure time: 9.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)