[English] 日本語
 Yorodumi
Yorodumi- EMDB-23601: Cryo-EM structure of CasPhi-2 (Cas12j) bound to crRNA and Phospho... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23601 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of CasPhi-2 (Cas12j) bound to crRNA and Phosphorothioate-DNA | |||||||||
 Map data Map data | LocSpiral map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / CasPhi / Cas12j / Nuclease / R-loop / crRNA / PAM / RNP / Complex / VIRAL PROTEIN-RNA-DNA / VIRAL PROTEIN-RNA-DNA complex | |||||||||
| Biological species |  Biggievirus Mos11 Biggievirus Mos11 | |||||||||
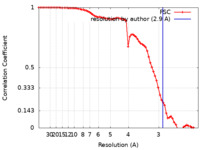
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Pausch P / Soczek K | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2021 Journal: Nat Struct Mol Biol / Year: 2021Title: DNA interference states of the hypercompact CRISPR-CasΦ effector. Authors: Patrick Pausch / Katarzyna M Soczek / Dominik A Herbst / Connor A Tsuchida / Basem Al-Shayeb / Jillian F Banfield / Eva Nogales / Jennifer A Doudna /  Abstract: CRISPR-CasΦ, a small RNA-guided enzyme found uniquely in bacteriophages, achieves programmable DNA cutting as well as genome editing. To investigate how the hypercompact enzyme recognizes and ...CRISPR-CasΦ, a small RNA-guided enzyme found uniquely in bacteriophages, achieves programmable DNA cutting as well as genome editing. To investigate how the hypercompact enzyme recognizes and cleaves double-stranded DNA, we determined cryo-EM structures of CasΦ (Cas12j) in pre- and post-DNA-binding states. The structures reveal a streamlined protein architecture that tightly encircles the CRISPR RNA and DNA target to capture, unwind and cleave DNA. Comparison of the pre- and post-DNA-binding states reveals how the protein rearranges for DNA cleavage upon target recognition. On the basis of these structures, we created and tested mutant forms of CasΦ that cut DNA up to 20-fold faster relative to wild type, showing how this system may be naturally attenuated to improve the fidelity of DNA interference. The structural and mechanistic insights into how CasΦ binds and cleaves DNA should allow for protein engineering for both in vitro diagnostics and genome editing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23601.map.gz emd_23601.map.gz | 5.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23601-v30.xml emd-23601-v30.xml emd-23601.xml emd-23601.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23601_fsc.xml emd_23601_fsc.xml | 7.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_23601.png emd_23601.png | 58.7 KB | ||
| Masks |  emd_23601_msk_1.map emd_23601_msk_1.map | 32.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23601.cif.gz emd-23601.cif.gz | 6.6 KB | ||
| Others |  emd_23601_additional_1.map.gz emd_23601_additional_1.map.gz emd_23601_half_map_1.map.gz emd_23601_half_map_1.map.gz emd_23601_half_map_2.map.gz emd_23601_half_map_2.map.gz | 16.4 MB 30.1 MB 30.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23601 http://ftp.pdbj.org/pub/emdb/structures/EMD-23601 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23601 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23601 | HTTPS FTP |
-Validation report
| Summary document |  emd_23601_validation.pdf.gz emd_23601_validation.pdf.gz | 996.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23601_full_validation.pdf.gz emd_23601_full_validation.pdf.gz | 995.9 KB | Display | |
| Data in XML |  emd_23601_validation.xml.gz emd_23601_validation.xml.gz | 14.6 KB | Display | |
| Data in CIF |  emd_23601_validation.cif.gz emd_23601_validation.cif.gz | 18.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23601 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23601 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23601 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23601 | HTTPS FTP |
-Related structure data
| Related structure data |  7lytMC  7lysC  7m5oC C: citing same article ( M: atomic model generated by this map |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23601.map.gz / Format: CCP4 / Size: 32.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23601.map.gz / Format: CCP4 / Size: 32.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LocSpiral map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.115 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23601_msk_1.map emd_23601_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: cryoSPARC map
| File | emd_23601_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoSPARC map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_23601_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_23601_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM map of CasPhi bound to crRNA and phosphorothioate-DNA in ...
| Entire | Name: Cryo-EM map of CasPhi bound to crRNA and phosphorothioate-DNA in the presence of the nuclease magnesium cofactor |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM map of CasPhi bound to crRNA and phosphorothioate-DNA in ...
| Supramolecule | Name: Cryo-EM map of CasPhi bound to crRNA and phosphorothioate-DNA in the presence of the nuclease magnesium cofactor type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Biggievirus Mos11 Biggievirus Mos11 |
| Molecular weight | Theoretical: 128 KDa |
-Macromolecule #1: CasPhi
| Macromolecule | Name: CasPhi / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Biggievirus Mos11 Biggievirus Mos11 |
| Molecular weight | Theoretical: 86.127188 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPKPAVESEF SKVLKKHFPG ERFRSSYMKR GGKILAAQGE EAVVAYLQGK SEEEPPNFQP PAKCHVVTKS RDFAEWPIMK ASEAIQRYI YALSTTERAA CKPGKSSESH AAWFAATGVS NHGYSHVQGL NLIFDHTLGR YDGVLKKVQL RNEKARARLE S INASRADE ...String: MPKPAVESEF SKVLKKHFPG ERFRSSYMKR GGKILAAQGE EAVVAYLQGK SEEEPPNFQP PAKCHVVTKS RDFAEWPIMK ASEAIQRYI YALSTTERAA CKPGKSSESH AAWFAATGVS NHGYSHVQGL NLIFDHTLGR YDGVLKKVQL RNEKARARLE S INASRADE GLPEIKAEEE EVATNETGHL LQPPGINPSF YVYQTISPQA YRPRDEIVLP PEYAGYVRDP NAPIPLGVVR NR CDIQKGC PGYIPEWQRE AGTAISPKTG KAVTVPGLSP KKNKRMRRYW RSEKEKAQDA LLVTVRIGTD WVVIDVRGLL RNA RWRTIA PKDISLNALL DLFTGDPVID VRRNIVTFTY TLDACGTYAR KWTLKGKQTK ATLDKLTATQ TVALVAIDLG QTNP ISAGI SRVTQENGAL QCEPLDRFTL PDDLLKDISA YRIAWDRNEE ELRARSVEAL PEAQQAEVRA LDGVSKETAR TQLCA DFGL DPKRLPWDKM SSNTTFISEA LLSNSVSRDQ VFFTPAPKKG AKKKAPVEVM RKDRTWARAY KPRLSVEAQK LKNEAL WAL KRTSPEYLKL SRRKEELCRR SINYVIEKTR RRTQCQIVIP VIEDLNVRFF HGSGKRLPGW DNFFTAKKEN RWFIQGL HK AFSDLRTHRS FYVFEVRPER TSITCPKCGH CEVGNRDGEA FQCLSCGKTC NADLDVATHN LTQVALTGKT MPKREEPR D AQGTAPARKT KKASKSKAPP AEREDQTPAQ EPSQTSHHHH HH |
-Macromolecule #2: crRNA
| Macromolecule | Name: crRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Biggievirus Mos11 Biggievirus Mos11 |
| Molecular weight | Theoretical: 14.481651 KDa |
| Sequence | String: CAACGAUUGC CCCUCACGAG GGGACAGCUG GUAAUGGGAU ACCUU |
-Macromolecule #3: TS-DNA
| Macromolecule | Name: TS-DNA / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Biggievirus Mos11 Biggievirus Mos11 |
| Molecular weight | Theoretical: 13.615714 KDa |
| Sequence | String: (DC)(DG)(DG)(DA)(DG)(DC)(DG)(DG)(DA)(DG) (DG)(DG)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DA) (DT)(DC)(DC)(DC)(DA)(DT)(DT)(DA)(DC) (DC)(DA)(DG)(DC)(DT)(DT)(DA)(DA)(DC)(DT) (DA) (DC)(DG)(DC)(DG) |
-Macromolecule #4: NTS-DNA*
| Macromolecule | Name: NTS-DNA* / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Biggievirus Mos11 Biggievirus Mos11 |
| Molecular weight | Theoretical: 13.072435 KDa |
| Sequence | String: (DC)(DG)(DC)(DG)(DT)(DA)(DG)(DT)(DT)(DA) (DT)(DC)(DG)(DA)(DC)(DC)(DA)(DT)(DT)(DA) (SC)(SC)(SC)(N)(N)(N)(DG)(DG)(DG) (DC)(DC)(DA)(DC)(DC)(DC)(DT)(DC)(DC)(DG) (DC) (DT)(DC)(DC)(DG) |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY |
| Vitrification | Cryogen name: NITROGEN / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)