[English] 日本語
 Yorodumi
Yorodumi- EMDB-2352: Structure of the Bunyamwera virus glycoprotein spike determined b... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2352 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
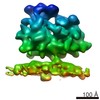
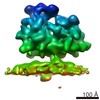
| Title | Structure of the Bunyamwera virus glycoprotein spike determined by electron cryo-tomography and sub-volume averaging | |||||||||
 Map data Map data | Sub-tomogram average of Bunyamwera virus glycoprotein spike | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bunyavirus / viral glycoprotein / reconstruction / viral fusion / orthobunyavirus | |||||||||
| Biological species |  Bunyamwera virus Bunyamwera virus | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 30.0 Å | |||||||||
 Authors Authors | Bowden TA / Bitto D / McLees A / Yeromonahos C / Elliott RM / Huiskonen JT | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2013 Journal: PLoS Pathog / Year: 2013Title: Orthobunyavirus ultrastructure and the curious tripodal glycoprotein spike. Authors: Thomas A Bowden / David Bitto / Angela McLees / Christelle Yeromonahos / Richard M Elliott / Juha T Huiskonen /  Abstract: The genus Orthobunyavirus within the family Bunyaviridae constitutes an expanding group of emerging viruses, which threaten human and animal health. Despite the medical importance, little is known ...The genus Orthobunyavirus within the family Bunyaviridae constitutes an expanding group of emerging viruses, which threaten human and animal health. Despite the medical importance, little is known about orthobunyavirus structure, a prerequisite for understanding virus assembly and entry. Here, using electron cryo-tomography, we report the ultrastructure of Bunyamwera virus, the prototypic member of this genus. Whilst Bunyamwera virions are pleomorphic in shape, they display a locally ordered lattice of glycoprotein spikes. Each spike protrudes 18 nm from the viral membrane and becomes disordered upon introduction to an acidic environment. Using sub-tomogram averaging, we derived a three-dimensional model of the trimeric pre-fusion glycoprotein spike to 3-nm resolution. The glycoprotein spike consists mainly of the putative class-II fusion glycoprotein and exhibits a unique tripod-like arrangement. Protein-protein contacts between neighbouring spikes occur at membrane-proximal regions and intra-spike contacts at membrane-distal regions. This trimeric assembly deviates from previously observed fusion glycoprotein arrangements, suggesting a greater than anticipated repertoire of viral fusion glycoprotein oligomerization. Our study provides evidence of a pH-dependent conformational change that occurs during orthobunyaviral entry into host cells and a blueprint for the structure of this group of emerging pathogens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2352.map.gz emd_2352.map.gz | 3.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2352-v30.xml emd-2352-v30.xml emd-2352.xml emd-2352.xml | 9.2 KB 9.2 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2352.png EMD-2352.png | 129.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2352 http://ftp.pdbj.org/pub/emdb/structures/EMD-2352 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2352 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2352 | HTTPS FTP |
-Validation report
| Summary document |  emd_2352_validation.pdf.gz emd_2352_validation.pdf.gz | 211.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2352_full_validation.pdf.gz emd_2352_full_validation.pdf.gz | 211 KB | Display | |
| Data in XML |  emd_2352_validation.xml.gz emd_2352_validation.xml.gz | 5.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2352 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2352 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2352 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2352 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2352.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2352.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sub-tomogram average of Bunyamwera virus glycoprotein spike | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Purified Bunyamwera virus particles
| Entire | Name: Purified Bunyamwera virus particles |
|---|---|
| Components |
|
-Supramolecule #1000: Purified Bunyamwera virus particles
| Supramolecule | Name: Purified Bunyamwera virus particles / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Macromolecule #1: Bunyamwera virus glycoprotein Gn-Gc membrane complex
| Macromolecule | Name: Bunyamwera virus glycoprotein Gn-Gc membrane complex / type: protein_or_peptide / ID: 1 / Name.synonym: Bunyamwera viral glycoprotein spike Details: One central trimeric spike surrounded by six neighbouring spikes Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Bunyamwera virus Bunyamwera virus |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 10 mM Tris pH 7.4, 100 mM NaCl |
|---|---|
| Grid | Details: C-flat CF-2/1/2C |
| Vitrification | Cryogen name: ETHANE-PROPANE MIXTURE / Chamber temperature: 103 K / Instrument: GATAN CRYOPLUNGE 3 / Method: Blot both sides for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 100 K / Max: 120 K / Average: 100 K |
| Specialist optics | Energy filter - Name: GIF2002 / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Details | Low dose tomography |
| Date | Mar 28, 2012 |
| Image recording | Category: CCD / Film or detector model: GENERIC CCD / Average electron dose: 100 e/Å2 Details: Thirteen single-axis tilt series were collected over an angular range of -60 to 60 at 3 degree increments. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 111111 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 4.5 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder: Liquid nitrogen cooled / Specimen holder model: OTHER / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | 2,000 subtomograms were selected using template matching in Jsubtomo |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 30.0 Å / Resolution method: OTHER / Software - Name: Jsubtomo Details: Final map was calculated by averaging the two half maps. A low pass filter to 30 A was applied. Number subtomograms used: 2000 |
| CTF correction | Details: Low pass filter was applied after the first zero of the CTF |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















