+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23093 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of YiiP-Fab complex in Holo state | |||||||||
 Map data Map data | locally sharpened map from cryosparc non-uniform refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Zinc transport / cation diffusion facilitator / holo state / inward-facing state / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationzinc efflux antiporter activity / cadmium ion transmembrane transporter activity / ferrous iron transmembrane transporter activity / intracellular zinc ion homeostasis / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Shewanella oneidensis (bacteria) / Shewanella oneidensis (bacteria) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
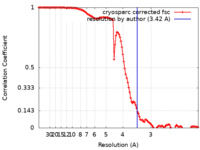
| Method | single particle reconstruction / cryo EM / Resolution: 3.42 Å | |||||||||
 Authors Authors | Lopez-Redondo ML / Fan S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Gen Physiol / Year: 2021 Journal: J Gen Physiol / Year: 2021Title: Zinc binding alters the conformational dynamics and drives the transport cycle of the cation diffusion facilitator YiiP. Authors: Maria Lopez-Redondo / Shujie Fan / Akiko Koide / Shohei Koide / Oliver Beckstein / David L Stokes /  Abstract: YiiP is a secondary transporter that couples Zn2+ transport to the proton motive force. Structural studies of YiiP from prokaryotes and Znt8 from humans have revealed three different Zn2+ sites and a ...YiiP is a secondary transporter that couples Zn2+ transport to the proton motive force. Structural studies of YiiP from prokaryotes and Znt8 from humans have revealed three different Zn2+ sites and a conserved homodimeric architecture. These structures define the inward-facing and outward-facing states that characterize the archetypal alternating access mechanism of transport. To study the effects of Zn2+ binding on the conformational transition, we use cryo-EM together with molecular dynamics simulation to compare structures of YiiP from Shewanella oneidensis in the presence and absence of Zn2+. To enable single-particle cryo-EM, we used a phage-display library to develop a Fab antibody fragment with high affinity for YiiP, thus producing a YiiP/Fab complex. To perform MD simulations, we developed a nonbonded dummy model for Zn2+ and validated its performance with known Zn2+-binding proteins. Using these tools, we find that, in the presence of Zn2+, YiiP adopts an inward-facing conformation consistent with that previously seen in tubular crystals. After removal of Zn2+ with high-affinity chelators, YiiP exhibits enhanced flexibility and adopts a novel conformation that appears to be intermediate between inward-facing and outward-facing states. This conformation involves closure of a hydrophobic gate that has been postulated to control access to the primary transport site. Comparison of several independent cryo-EM maps suggests that the transition from the inward-facing state is controlled by occupancy of a secondary Zn2+ site at the cytoplasmic membrane interface. This work enhances our understanding of individual Zn2+ binding sites and their role in the conformational dynamics that govern the transport cycle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23093.map.gz emd_23093.map.gz | 129.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23093-v30.xml emd-23093-v30.xml emd-23093.xml emd-23093.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23093_fsc.xml emd_23093_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_23093.png emd_23093.png | 159.9 KB | ||
| Masks |  emd_23093_msk_1.map emd_23093_msk_1.map | 137.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23093.cif.gz emd-23093.cif.gz | 7.1 KB | ||
| Others |  emd_23093_half_map_1.map.gz emd_23093_half_map_1.map.gz emd_23093_half_map_2.map.gz emd_23093_half_map_2.map.gz | 126.9 MB 126.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23093 http://ftp.pdbj.org/pub/emdb/structures/EMD-23093 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23093 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23093 | HTTPS FTP |
-Validation report
| Summary document |  emd_23093_validation.pdf.gz emd_23093_validation.pdf.gz | 873.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23093_full_validation.pdf.gz emd_23093_full_validation.pdf.gz | 873.1 KB | Display | |
| Data in XML |  emd_23093_validation.xml.gz emd_23093_validation.xml.gz | 19.5 KB | Display | |
| Data in CIF |  emd_23093_validation.cif.gz emd_23093_validation.cif.gz | 25.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23093 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23093 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23093 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23093 | HTTPS FTP |
-Related structure data
| Related structure data |  7kzzMC  7kzxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23093.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23093.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | locally sharpened map from cryosparc non-uniform refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.048 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23093_msk_1.map emd_23093_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half-map from cryosparc non-uniform refinement
| File | emd_23093_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map from cryosparc non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half-map from cryosparc non-uniform refinement
| File | emd_23093_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map from cryosparc non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : YiiP_SO in complex with Fab2R
| Entire | Name: YiiP_SO in complex with Fab2R |
|---|---|
| Components |
|
-Supramolecule #1: YiiP_SO in complex with Fab2R
| Supramolecule | Name: YiiP_SO in complex with Fab2R / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: YiiP-Fab complex purified in decyl maltopyranoside (DM) detergent |
|---|---|
| Molecular weight | Theoretical: 25 KDa |
-Supramolecule #2: YiiP
| Supramolecule | Name: YiiP / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Shewanella oneidensis (bacteria) Shewanella oneidensis (bacteria) |
-Supramolecule #3: Fab2r light chain
| Supramolecule | Name: Fab2r light chain / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: Fab2r heavy chain
| Supramolecule | Name: Fab2r heavy chain / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Cadmium and zinc efflux pump FieF
| Macromolecule | Name: Cadmium and zinc efflux pump FieF / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shewanella oneidensis (bacteria) Shewanella oneidensis (bacteria) |
| Molecular weight | Theoretical: 32.485211 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTQTSQYDFW VKLASRASVA TALTLITIKL LAWLYSGSAS MLASLTDSFA DTLASIINFI AIRYAIVPAD HDHRYGHGKA EPLAALAQS AFIMGSAFLL LFYGGERLLN PSPVENATLG VVVSVVAIVL TLALVLLQKR ALAATNSTVV EADSLHYKSD L FLNAAVLL ...String: MTQTSQYDFW VKLASRASVA TALTLITIKL LAWLYSGSAS MLASLTDSFA DTLASIINFI AIRYAIVPAD HDHRYGHGKA EPLAALAQS AFIMGSAFLL LFYGGERLLN PSPVENATLG VVVSVVAIVL TLALVLLQKR ALAATNSTVV EADSLHYKSD L FLNAAVLL ALVLSQYGWW WADGLFAVLI ACYIGQQAFD LGYRSIQALL DRELDEDTRQ RIKLIAKEDP RVLGLHDLRT RQ AGKTVFI QFHLELDGNL SLNEAHSITD TTGLRVKAAF EDAEVIIHQD PVQVEPTTQ UniProtKB: Cation-efflux pump FieF |
-Macromolecule #2: Fab2R light chain
| Macromolecule | Name: Fab2R light chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.580242 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQIWSWPLIT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQIWSWPLIT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Macromolecule #3: Fab2R heavy chain
| Macromolecule | Name: Fab2R heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.406352 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF TIYSSSIHWV RQAPGKGLEW VASIYSSSGS TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARQSYSGLSP RRHWSYGAMD YWGQGTLVTV FNQIKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF TIYSSSIHWV RQAPGKGLEW VASIYSSSGS TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARQSYSGLSP RRHWSYGAMD YWGQGTLVTV FNQIKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSCDKTHT |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 8 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 5 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.02 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 3 uL of protein mixture applied to grid. Blot time 4 seconds, blot force 0.. | |||||||||||||||
| Details | YiiP-Fab2R complex purified through SEC with DM, the same day as grid freezing. Main Peak fraction was used for freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 77.0 K / Max: 77.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 15 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Number grids imaged: 1 / Number real images: 3674 / Average exposure time: 9.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)