[English] 日本語
 Yorodumi
Yorodumi- EMDB-21945: Kif14[391-743] - ADP-AlFx open state class in complex with a micr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21945 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Kif14[391-743] - ADP-AlFx open state class in complex with a microtubule | |||||||||||||||||||||||||||
 Map data Map data | Main map. Locally refined single unit. Not helically averaged. | |||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | KIF14 / kinesin / motility / microtubule / tubulin / MOTOR PROTEIN | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcerebellar granular layer structural organization / regulation of cell maturation / RHO GTPases activate CIT / cerebellar Purkinje cell layer structural organization / negative regulation of integrin activation / RND2 GTPase cycle / RND1 GTPase cycle / cell proliferation in forebrain / regulation of Rap protein signal transduction / cerebellar cortex development ...cerebellar granular layer structural organization / regulation of cell maturation / RHO GTPases activate CIT / cerebellar Purkinje cell layer structural organization / negative regulation of integrin activation / RND2 GTPase cycle / RND1 GTPase cycle / cell proliferation in forebrain / regulation of Rap protein signal transduction / cerebellar cortex development / olfactory bulb development / Flemming body / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Mitotic Prometaphase / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / EML4 and NUDC in mitotic spindle formation / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / PKR-mediated signaling / Separation of Sister Chromatids / The role of GTSE1 in G2/M progression after G2 checkpoint / Aggrephagy / plus-end-directed microtubule motor activity / activation of protein kinase activity / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / MHC class II antigen presentation / Recruitment of NuMA to mitotic centrosomes / COPI-mediated anterograde transport / regulation of myelination / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / microtubule-based movement / mitotic metaphase chromosome alignment / positive regulation of cytokinesis / regulation of G1/S transition of mitotic cell cycle / spindle midzone / regulation of cell adhesion / microtubule-based process / regulation of neuron apoptotic process / regulation of G2/M transition of mitotic cell cycle / tubulin binding / substrate adhesion-dependent cell spreading / regulation of cell migration / hippocampus development / PDZ domain binding / regulation of cell growth / establishment of protein localization / cerebral cortex development / structural constituent of cytoskeleton / microtubule cytoskeleton organization / mitotic cell cycle / microtubule cytoskeleton / midbody / microtubule binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / negative regulation of neuron apoptotic process / proteasome-mediated ubiquitin-dependent protein catabolic process / microtubule / cell division / GTPase activity / positive regulation of cell population proliferation / negative regulation of apoptotic process / protein kinase binding / GTP binding / ATP hydrolysis activity / ATP binding / metal ion binding / nucleus / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |   | |||||||||||||||||||||||||||
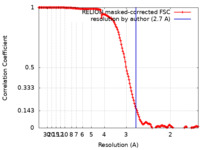
| Method | helical reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||||||||||||||||||||
 Authors Authors | Benoit MPMH / Asenjo AB | |||||||||||||||||||||||||||
| Funding support |  United States, United States,  Canada, 8 items Canada, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis of mechano-chemical coupling by the mitotic kinesin KIF14. Authors: Matthieu P M H Benoit / Ana B Asenjo / Mohammadjavad Paydar / Sabin Dhakal / Benjamin H Kwok / Hernando Sosa /   Abstract: KIF14 is a mitotic kinesin whose malfunction is associated with cerebral and renal developmental defects and several cancers. Like other kinesins, KIF14 couples ATP hydrolysis and microtubule binding ...KIF14 is a mitotic kinesin whose malfunction is associated with cerebral and renal developmental defects and several cancers. Like other kinesins, KIF14 couples ATP hydrolysis and microtubule binding to the generation of mechanical work, but the coupling mechanism between these processes is still not fully clear. Here we report 20 high-resolution (2.7-3.9 Å) cryo-electron microscopy KIF14-microtubule structures with complementary functional assays. Analysis procedures were implemented to separate coexisting conformations of microtubule-bound monomeric and dimeric KIF14 constructs. The data provide a comprehensive view of the microtubule and nucleotide induced KIF14 conformational changes. It shows that: 1) microtubule binding, the nucleotide species, and the neck-linker domain govern the transition between three major conformations of the motor domain; 2) an undocked neck-linker prevents the nucleotide-binding pocket to fully close and dampens ATP hydrolysis; 3) 13 neck-linker residues are required to assume a stable docked conformation; 4) the neck-linker position controls the hydrolysis rather than the nucleotide binding step; 5) the two motor domains of KIF14 dimers adopt distinct conformations when bound to the microtubule; and 6) the formation of the two-heads-bound-state introduces structural changes in both motor domains of KIF14 dimers. These observations provide the structural basis for a coordinated chemo-mechanical kinesin translocation model. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21945.map.gz emd_21945.map.gz | 7.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21945-v30.xml emd-21945-v30.xml emd-21945.xml emd-21945.xml | 34.9 KB 34.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21945_fsc.xml emd_21945_fsc.xml | 14.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_21945.png emd_21945.png | 159.6 KB | ||
| Masks |  emd_21945_msk_1.map emd_21945_msk_1.map emd_21945_msk_2.map emd_21945_msk_2.map emd_21945_msk_3.map emd_21945_msk_3.map emd_21945_msk_4.map emd_21945_msk_4.map emd_21945_msk_5.map emd_21945_msk_5.map emd_21945_msk_6.map emd_21945_msk_6.map | 1.1 GB 274.6 MB 1.1 GB 1.1 GB 274.6 MB 274.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-21945.cif.gz emd-21945.cif.gz | 8.3 KB | ||
| Others |  emd_21945_additional_1.map.gz emd_21945_additional_1.map.gz emd_21945_additional_2.map.gz emd_21945_additional_2.map.gz emd_21945_additional_3.map.gz emd_21945_additional_3.map.gz emd_21945_half_map_1.map.gz emd_21945_half_map_1.map.gz emd_21945_half_map_2.map.gz emd_21945_half_map_2.map.gz | 822.6 MB 213.8 MB 822.7 MB 218.7 MB 218.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21945 http://ftp.pdbj.org/pub/emdb/structures/EMD-21945 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21945 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21945 | HTTPS FTP |
-Validation report
| Summary document |  emd_21945_validation.pdf.gz emd_21945_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21945_full_validation.pdf.gz emd_21945_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_21945_validation.xml.gz emd_21945_validation.xml.gz | 22 KB | Display | |
| Data in CIF |  emd_21945_validation.cif.gz emd_21945_validation.cif.gz | 29.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21945 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21945 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21945 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21945 | HTTPS FTP |
-Related structure data
| Related structure data |  6wwrMC  6wweC  6wwfC  6wwgC  6wwhC  6wwiC  6wwjC  6wwkC  6wwlC  6wwmC  6wwnC  6wwoC  6wwpC  6wwqC  6wwsC  6wwtC  6wwuC  6wwvC  7lvqC  7lvrC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21945.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21945.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map. Locally refined single unit. Not helically averaged. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
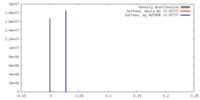
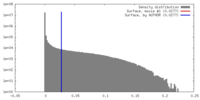
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Mask #1
+Mask #2
+Mask #3
+Mask #4
+Mask #5
+Mask #6
+Additional map: Helical reconstruction half map 2 (gold standard).
+Additional map: Helical reconstruction.
+Additional map: Helical reconstruction half map 1 (gold standard).
+Half map: Half map 1 (gold standard). Locally refined single...
+Half map: Half map 2 (gold standard). Locally refined single...
- Sample components
Sample components
+Entire : Kif14[391-743] - ADP-AlFx in complex with a microtubule
+Supramolecule #1: Kif14[391-743] - ADP-AlFx in complex with a microtubule
+Supramolecule #2: KIF14[391-743] - ADP-AlFx
+Supramolecule #3: Microtubule
+Macromolecule #1: Tubulin alpha-1B chain
+Macromolecule #2: Tubulin beta-2B chain
+Macromolecule #3: Kinesin-like protein KIF14
+Macromolecule #4: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #5: MAGNESIUM ION
+Macromolecule #6: GUANOSINE-5'-DIPHOSPHATE
+Macromolecule #7: TAXOL
+Macromolecule #8: ADENOSINE-5'-DIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 Component:
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 200 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING | ||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Average electron dose: 70.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 60606 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6wwr: |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)