[English] 日本語
 Yorodumi
Yorodumi- EMDB-19981: Cryo-EM structure of the full-length Pseudomonas aeruginosa bacte... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the full-length Pseudomonas aeruginosa bacteriophytochrome in its Pr state | |||||||||
 Map data Map data | Composite map generated with phenix combine maps. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Photosensor / Photoreceptor / Phytochrome / Bacterial protein / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationosmosensory signaling via phosphorelay pathway / detection of visible light / phosphorelay response regulator activity / protein kinase activator activity / histidine kinase / phosphorelay sensor kinase activity / photoreceptor activity / regulation of DNA-templated transcription / ATP binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
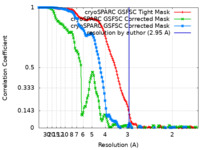
| Method | single particle reconstruction / cryo EM / Resolution: 2.95 Å | |||||||||
 Authors Authors | Bodizs S / Westenhoff S | |||||||||
| Funding support |  Sweden, 1 items Sweden, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Cryo-EM structures of a bathy phytochrome histidine kinase reveal a unique light-dependent activation mechanism. Authors: Szabolcs Bódizs / Petra Mészáros / Lukas Grunewald / Heikki Takala / Sebastian Westenhoff /   Abstract: Phytochromes are photoreceptor proteins in plants, fungi, and bacteria. They can adopt two photochromic states with differential biochemical responses. The structural changes transducing the signal ...Phytochromes are photoreceptor proteins in plants, fungi, and bacteria. They can adopt two photochromic states with differential biochemical responses. The structural changes transducing the signal from the chromophore to the biochemical output modules are poorly understood due to challenges in capturing structures of the dynamic, full-length protein. Here, we present cryoelectron microscopy (cryo-EM) structures of the phytochrome from Pseudomonas aeruginosa (PaBphP) in its resting (Pfr) and photoactivated (Pr) state. The kinase-active Pr state has an asymmetric, dimeric structure, whereas the kinase-inactive Pfr state opens up. This behavior is different from other known phytochromes and we explain it with the unusually short connection between the photosensory and output modules. Multiple sequence alignment of this region suggests evolutionary optimization for different modes of signal transduction in sensor proteins. The results establish a new mechanism for light-sensing by phytochrome histidine kinases and provide input for the design of optogenetic phytochrome variants. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19981.map.gz emd_19981.map.gz | 230.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19981-v30.xml emd-19981-v30.xml emd-19981.xml emd-19981.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19981_fsc.xml emd_19981_fsc.xml emd_19981_fsc_2.xml emd_19981_fsc_2.xml emd_19981_fsc_3.xml emd_19981_fsc_3.xml | 13.3 KB 13.3 KB 13.3 KB | Display Display Display |  FSC data file FSC data file |
| Images |  emd_19981.png emd_19981.png | 69.3 KB | ||
| Filedesc metadata |  emd-19981.cif.gz emd-19981.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19981 http://ftp.pdbj.org/pub/emdb/structures/EMD-19981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19981 | HTTPS FTP |
-Validation report
| Summary document |  emd_19981_validation.pdf.gz emd_19981_validation.pdf.gz | 495.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19981_full_validation.pdf.gz emd_19981_full_validation.pdf.gz | 495.5 KB | Display | |
| Data in XML |  emd_19981_validation.xml.gz emd_19981_validation.xml.gz | 14.2 KB | Display | |
| Data in CIF |  emd_19981_validation.cif.gz emd_19981_validation.cif.gz | 18.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19981 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19981 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19981 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19981 | HTTPS FTP |
-Related structure data
| Related structure data |  9eutMC  9euyC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19981.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19981.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map generated with phenix combine maps. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Homodimeric complex of the protein bacteriophytochrome containing...
| Entire | Name: Homodimeric complex of the protein bacteriophytochrome containing its cofactor biliverdin |
|---|---|
| Components |
|
-Supramolecule #1: Homodimeric complex of the protein bacteriophytochrome containing...
| Supramolecule | Name: Homodimeric complex of the protein bacteriophytochrome containing its cofactor biliverdin type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 160 KDa |
-Macromolecule #1: Bacteriophytochrome
| Macromolecule | Name: Bacteriophytochrome / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: histidine kinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 82.094867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTSITPVTLA NCEDEPIHVP GAIQPHGALV TLRADGMVLA ASENIQALLG FVASPGSYLT QEQVGPEVLR MLEEGLTGNG PWSNSVETR IGEHLFDVIG HSYKEVFYLE FEIRTADTLS ITSFTLNAQR IIAQVQLHND TASLLSNVTD ELRRMTGYDR V MAYRFRHD ...String: MTSITPVTLA NCEDEPIHVP GAIQPHGALV TLRADGMVLA ASENIQALLG FVASPGSYLT QEQVGPEVLR MLEEGLTGNG PWSNSVETR IGEHLFDVIG HSYKEVFYLE FEIRTADTLS ITSFTLNAQR IIAQVQLHND TASLLSNVTD ELRRMTGYDR V MAYRFRHD DSGEVVAESR REDLESYLGQ RYPASDIPAQ ARRLYIQNPI RLIADVAYTP MRVFPALNPE TNESFDLSYS VL RSVSPIH CEYLTNMGVR ASMSISIVVG GKLWGLFSCH HMSPKLIPYP VRMSFQIFSQ VCSAIVERLE QGRIAELLRV STE RRLALA RRARDADDLF GALAHPDDGI AALIPCDGAL VMLGGRTLSI RGDFERQAGN VLQRLQRDPE RDIYHTDNWP QPSE DSPDG GDCCGVLAIR FHRQESGWIF WFRHEEVHRI RWGGKPEKLL TIGPSGPRLT PRGSFEAWEE VVRGHSTPWS ETDLA IAEK LRLDLMELCL NHAAEVDRMR QRLIAVLGHD LRNPLQSISM AAALLSSSDT RTTELRQHIS ASSSRMERLV SQILDM SRL QSGIGLTVNP VDTDVSQLVR QIVCETDVAY PGLVIEIAID PQVRAVVDPD RYAQVAANLL SNARHHGLPG RPVLVTL TR QGDEVCLSVL NETSGLSEAQ LANLFEPFKR ESADNQRNRN GLGIGLYISQ AIAQAHQGRI DVDCRDDVIT FCLRLPVR Q AETGSSSLEH HHHHH UniProtKB: Bacteriophytochrome |
-Macromolecule #2: 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidan...
| Macromolecule | Name: 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene- ...Name: 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid type: ligand / ID: 2 / Number of copies: 2 / Formula: LBV |
|---|---|
| Molecular weight | Theoretical: 585.67 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
Details: 80 mM Tris, 10 mM MgCl2, 150 mM CH3CO2K, pH 7.8 | ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 0.04 kPa / Details: Current: 20 mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting for 5 s from both sides. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 2 / Number real images: 30336 / Average exposure time: 1.7 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 2.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Output model |  PDB-9eut: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)