+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | High resolution structure of FZD7 in inactive conformation | ||||||||||||||||||||||||
 Map data Map data | High resolution structure of inactive FZD7 | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | GPCPR / Frizzled / FZD7 / MEMBRANE PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of ectodermal cell fate specification / negative regulation of cardiac muscle cell differentiation / mesenchymal to epithelial transition / skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration / somatic stem cell division / Wnt receptor activity / non-canonical Wnt signaling pathway / positive regulation of epithelial cell proliferation involved in wound healing / Wnt-protein binding / WNT5:FZD7-mediated leishmania damping ...negative regulation of ectodermal cell fate specification / negative regulation of cardiac muscle cell differentiation / mesenchymal to epithelial transition / skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration / somatic stem cell division / Wnt receptor activity / non-canonical Wnt signaling pathway / positive regulation of epithelial cell proliferation involved in wound healing / Wnt-protein binding / WNT5:FZD7-mediated leishmania damping / frizzled binding / PCP/CE pathway / Class B/2 (Secretin family receptors) / negative regulation of cell-substrate adhesion / regulation of canonical Wnt signaling pathway / Wnt signaling pathway, planar cell polarity pathway / stem cell population maintenance / canonical Wnt signaling pathway / positive regulation of phosphorylation / cellular response to retinoic acid / phosphatidylinositol-4,5-bisphosphate binding / substrate adhesion-dependent cell spreading / Asymmetric localization of PCP proteins / PDZ domain binding / G protein-coupled receptor activity / positive regulation of JNK cascade / neuron differentiation / recycling endosome membrane / T cell differentiation in thymus / positive regulation of MAPK cascade / intracellular membrane-bounded organelle / regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.9 Å | ||||||||||||||||||||||||
 Authors Authors | Bous J / Kinsolving J / Gratz L / Scharf MM / Voss J / Selcuk B / Adebali O / Schulte G | ||||||||||||||||||||||||
| Funding support |  Sweden, Sweden,  Germany, 7 items Germany, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: high-resolution structural characterization of FZD7 unravels the importance of water network and cholesterol for class F GPCRs dynamic and activation Authors: Bous J / Kinsolving J / Gratz L / Scharf MM / Voss J / Selcuk B / Adebali O / Schulte G | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19881.map.gz emd_19881.map.gz | 188.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19881-v30.xml emd-19881-v30.xml emd-19881.xml emd-19881.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19881_fsc.xml emd_19881_fsc.xml | 12.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_19881.png emd_19881.png | 186 KB | ||
| Masks |  emd_19881_msk_1.map emd_19881_msk_1.map | 199.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19881.cif.gz emd-19881.cif.gz | 6.7 KB | ||
| Others |  emd_19881_half_map_1.map.gz emd_19881_half_map_1.map.gz emd_19881_half_map_2.map.gz emd_19881_half_map_2.map.gz | 185.2 MB 185.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19881 http://ftp.pdbj.org/pub/emdb/structures/EMD-19881 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19881 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19881 | HTTPS FTP |
-Validation report
| Summary document |  emd_19881_validation.pdf.gz emd_19881_validation.pdf.gz | 1004.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19881_full_validation.pdf.gz emd_19881_full_validation.pdf.gz | 1004.4 KB | Display | |
| Data in XML |  emd_19881_validation.xml.gz emd_19881_validation.xml.gz | 20.9 KB | Display | |
| Data in CIF |  emd_19881_validation.cif.gz emd_19881_validation.cif.gz | 27 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19881 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19881 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19881 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19881 | HTTPS FTP |
-Related structure data
| Related structure data |  9epoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19881.map.gz / Format: CCP4 / Size: 199.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19881.map.gz / Format: CCP4 / Size: 199.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | High resolution structure of inactive FZD7 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.6768 Å | ||||||||||||||||||||||||||||||||||||
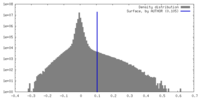
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19881_msk_1.map emd_19881_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
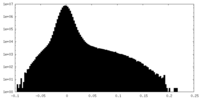
| Density Histograms |
-Half map: High resolution structure of inactive FZD7 (halph map 1)
| File | emd_19881_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | High resolution structure of inactive FZD7 (halph map 1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: High resolution structure of inactive FZD7 (half map 2)
| File | emd_19881_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | High resolution structure of inactive FZD7 (half map 2) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : FZD7 anti-parallel Dimer
| Entire | Name: FZD7 anti-parallel Dimer |
|---|---|
| Components |
|
-Supramolecule #1: FZD7 anti-parallel Dimer
| Supramolecule | Name: FZD7 anti-parallel Dimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 134788 kDa/nm |
-Macromolecule #1: Frizzled-7
| Macromolecule | Name: Frizzled-7 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.460859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDQPYHGEK GISVPDHGFC QPISIPLCTD IAYNQTILPN LLGHTNQEDA GLEVHQFYPL VKVQCSPEL RFFLCSMYAP VCTVLDQAIP PCRSLCERAR QGCEALMNKF GFQWPERLRC ENFPVHGAGE ICVGQNTSDG S GGPGGGPT ...String: MKTIIALSYI FCLVFADYKD DDDQPYHGEK GISVPDHGFC QPISIPLCTD IAYNQTILPN LLGHTNQEDA GLEVHQFYPL VKVQCSPEL RFFLCSMYAP VCTVLDQAIP PCRSLCERAR QGCEALMNKF GFQWPERLRC ENFPVHGAGE ICVGQNTSDG S GGPGGGPT AYPTAPYLPD LPFTALPPGA SDGRGRPAFP FSCPRQLKVP PYLGYRFLGE RDCGAPCEPG RANGLMYFKE EE RRFARLW VGVWSVLCCA STLFTVLTYL VDMRRFSYPE RPIIFLSGCY FMVAVAHVAG FLLEDRAVCV ERFSDDGYRT VAQ GTKKEG CTILFMVLYF FGMASSIWWV ILSLTWFLAA GMKWGHEAIE ANSQYFHLAA WAVPAVKTIT ILAMGQVDGD LLSG VCYVG LSSVDALRGF VLAPLFVYLF IGTSFLLAGF VSLFRIRTIM KHDGTKTEKL EKLMVRIGVF SVLYTVPATI VLACY FYEQ AFREHWERTW LLQTCKSYAV PCPPGHFPPM SPDFTVFMIK YLMTMIVGIT TGFWIWSGKT LQSWRRFYHR LSHSSK GET AVSRLEVLFQ GPWSHPQFEK GGGSGGGSGG GSWSHPQFEK UniProtKB: Frizzled-7 |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 4 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #3: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 3 / Number of copies: 2 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 94 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.08 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 21081 / Average exposure time: 3.5 sec. / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)