+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
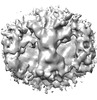
| Title | COPII inner coat on vesicles | |||||||||||||||
 Map data Map data | Map. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | COPII / coat / PROTEIN TRANSPORT | |||||||||||||||
| Biological species |  | |||||||||||||||
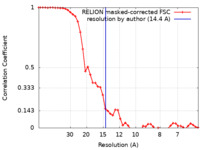
| Method | subtomogram averaging / cryo EM / Resolution: 14.4 Å | |||||||||||||||
 Authors Authors | Zanetti G / Pyle E | |||||||||||||||
| Funding support | European Union,  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-electron tomography reveals how COPII assembles on cargo-containing membranes Authors: Zanetti G / Pyle E | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19417.map.gz emd_19417.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19417-v30.xml emd-19417-v30.xml emd-19417.xml emd-19417.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19417_fsc.xml emd_19417_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_19417.png emd_19417.png | 108.3 KB | ||
| Masks |  emd_19417_msk_1.map emd_19417_msk_1.map | 8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19417.cif.gz emd-19417.cif.gz | 6.3 KB | ||
| Others |  emd_19417_half_map_1.map.gz emd_19417_half_map_1.map.gz emd_19417_half_map_2.map.gz emd_19417_half_map_2.map.gz | 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19417 http://ftp.pdbj.org/pub/emdb/structures/EMD-19417 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19417 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19417 | HTTPS FTP |
-Validation report
| Summary document |  emd_19417_validation.pdf.gz emd_19417_validation.pdf.gz | 873.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19417_full_validation.pdf.gz emd_19417_full_validation.pdf.gz | 873.2 KB | Display | |
| Data in XML |  emd_19417_validation.xml.gz emd_19417_validation.xml.gz | 10.4 KB | Display | |
| Data in CIF |  emd_19417_validation.cif.gz emd_19417_validation.cif.gz | 13.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19417 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19417 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19417 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19417 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19417.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19417.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.052 Å | ||||||||||||||||||||||||||||||||||||
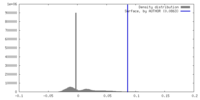
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19417_msk_1.map emd_19417_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
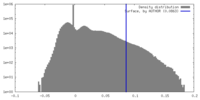
| Density Histograms |
-Half map: Half map 2.
| File | emd_19417_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
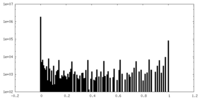
| Density Histograms |
-Half map: Half map 1.
| File | emd_19417_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
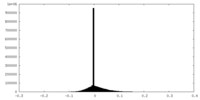
| Density Histograms |
- Sample components
Sample components
-Entire : COPII inner coat on vesicles
| Entire | Name: COPII inner coat on vesicles |
|---|---|
| Components |
|
-Supramolecule #1: COPII inner coat on vesicles
| Supramolecule | Name: COPII inner coat on vesicles / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: COPII inner coat on vesicles formed from native-like membranes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 210 KDa |
-Macromolecule #1: Sec24
| Macromolecule | Name: Sec24 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MSHHKKRVYP QAQLQYGQN A TPLQQPAQ FM PPQDPAA AGM SYGQMG MPPQ GAVPS MGQQQ FLTP AQEQLH QQI DQATTSM ND MHLHNVPL V DPNAYMQPQ VPVQMGTPLQ QQQQPMAAP A YGQPSAAM GQ NMRPMNQ LYP IDLLTE LPPP ITDLT ...String: MSHHKKRVYP QAQLQYGQN A TPLQQPAQ FM PPQDPAA AGM SYGQMG MPPQ GAVPS MGQQQ FLTP AQEQLH QQI DQATTSM ND MHLHNVPL V DPNAYMQPQ VPVQMGTPLQ QQQQPMAAP A YGQPSAAM GQ NMRPMNQ LYP IDLLTE LPPP ITDLT LPPPP LVIP PERMLV PSE LSNASPD YI RSTLNAVP K NSSLLKKSK LPFGLVIRPY QHLYDDIDP P PLNEDGLI VR CRRCRSY MNP FVTFIE QGRR WRCNF CRLAN DVPM QMDQSD PND PKSRYDR NE IKCAVMEY M APKEYTLRQ PPPATYCFLI DVSQSSIKS G LLATTINT LL QNLDSIP NHD ERTRIS ILCV DNAIH YFKIP LDSE NNEESA DQI NMMDIAD LE EPFLPRPN S MVVSLKACR QNIETLLTKI PQIFQSNLI T NFALGPAL KS AYHLIGG VGG KIIVVS GTLP NLGIG KLQRR NESG VVNTSK ETA QLLSCQD SF YKNFTIDC S KVQITVDLF LASEDYMDVA SLSNLSRFT A GQTHFYPG FS GKNPNDI VKF STEFAK HISM DFCME TVMRA RGST GLRMSR FYG HFFNRSS DL CAFSTMPR D QSYLFEVNV DESIMADYCY VQVAVLLSL N NSQRRIRI IT LAMPTTE SLA EVYASA DQLA IASFY NSKAV EKAL NSSLDD ARV LINKSVQ DI LATYKKEI V VSNTAGGAP LRLCANLRMF PLLMHSLTK H MAFRSGIV PS DHRASAL NNL ESLPLK YLIK NIYPD VYSLH DMAD EAGLPV QTE DGEATGT IV LPQPINAT S SLFERYGLY LIDNGNELFL WMGGDAVPA L VFDVFGTQ DI FDIPIGK QEI PVVENS EFNQ RVRNI INQLR NHDD VITYQS LYI VRGASLS EP VNHASARE V ATLRLWASS TLVEDKILNN ESYREFLQI M KARISK |
-Macromolecule #2: Sec23
| Macromolecule | Name: Sec23 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: DFETNEDING VRFTWNVFP S TRSDANSN VV PVGCLYT PLK EYDELN VAPY NPVVC SGPHC KSIL NPYCVI DPR NSSWSCP IC NSRNHLPP Q YTNLSQENM PLELQSTTIE YITNKPVTV P PIFFFVVD LT SETENLD SLK ESIITS LSLL PPNAL ...String: DFETNEDING VRFTWNVFP S TRSDANSN VV PVGCLYT PLK EYDELN VAPY NPVVC SGPHC KSIL NPYCVI DPR NSSWSCP IC NSRNHLPP Q YTNLSQENM PLELQSTTIE YITNKPVTV P PIFFFVVD LT SETENLD SLK ESIITS LSLL PPNAL IGLIT YGNV VQLHDL SSE TIDRCNV FR GDREYQLE A LTEMLTGQK PTGPGGAASH LPNAMNKVT P FSLNRFFL PL EQVEFKL NQL LENLSP DQWS VPAGH RPLRA TGSA LNIASL LLQ GCYKNIP AR IILFASGP G TVAPGLIVN SELKDPLRSH HDIDSDHAQ H YKKACKFY NQ IAQRVAA NGH TVDIFA GCYD QIGMS EMKQL TDST GGVLLL TDA FSTAIFK QS YLRLFAKD E EGYLKMAFN GNMAVKTSKD LKVQGLIGH A SAVKKTDA NN ISESEIG IGA TSTWKM ASLS PYHSY AIFFE IANT AANSNP MMS APGSADR PH LAYTQFIT T YQHSSGTNR IRVTTVANQL LPFGTPAIA A SFDQEAAA VL MARIAVH KAE TDDGAD VIRW LDRTL IKLCQ KYAD YNKDDP QSF RLAPNFS LY PQFTYYLR R SQFLSVFNN SPDETAFYRH IFTREDTTN S LIMIQPTL TS FSMEDDP QPV LLDSIS VKPN TILLL DTFFF ILIY HGEQIA QWR KAGYQDD PQ YADFKALL E EPKLEAAEL LVDRFPLPRF IDTEAGGSQ A RFLLSKLN PS DNYQDMA RGG STIVLT DDVS LQNFM THLQQ VAVS GQA |
-Macromolecule #3: Sar1
| Macromolecule | Name: Sar1 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MAGWDIFGWF RDVLASLGL W NKHGKLLF LG LDNAGKT TLL HMLKND RLAT LQPTW HPTSE ELAI GNIKFT TFD LGGHIQA RR LWKDYFPE V NGIVFLVDA ADPERFDEAR VELDALFNI A ELKDVPFV IL GNKIDAP NAV SEAELR SALG LLNTT ...String: MAGWDIFGWF RDVLASLGL W NKHGKLLF LG LDNAGKT TLL HMLKND RLAT LQPTW HPTSE ELAI GNIKFT TFD LGGHIQA RR LWKDYFPE V NGIVFLVDA ADPERFDEAR VELDALFNI A ELKDVPFV IL GNKIDAP NAV SEAELR SALG LLNTT GSQRI EGQR PVEVFM CSV VMRNGYL EA FQWLSQYI |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 3.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)