[English] 日本語
 Yorodumi
Yorodumi- EMDB-19222: in situ subtomogram average of MEF cell pre-60S ribosome in the s... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | in situ subtomogram average of MEF cell pre-60S ribosome in the state B | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ribosome 60S maturation subtomogram averaging / RIBOSOME | ||||||||||||
| Biological species |  | ||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 28.0 Å | ||||||||||||
 Authors Authors | Fedry J / Forster F | ||||||||||||
| Funding support | European Union,  Netherlands, 3 items Netherlands, 3 items
| ||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Visualization of translation reorganization upon persistent ribosome collision stress in mammalian cells. Authors: Juliette Fedry / Joana Silva / Mihajlo Vanevic / Stanley Fronik / Yves Mechulam / Emmanuelle Schmitt / Amédée des Georges / William James Faller / Friedrich Förster /     Abstract: Aberrantly slow ribosomes incur collisions, a sentinel of stress that triggers quality control, signaling, and translation attenuation. Although each collision response has been studied in isolation, ...Aberrantly slow ribosomes incur collisions, a sentinel of stress that triggers quality control, signaling, and translation attenuation. Although each collision response has been studied in isolation, the net consequences of their collective actions in reshaping translation in cells is poorly understood. Here, we apply cryoelectron tomography to visualize the translation machinery in mammalian cells during persistent collision stress. We find that polysomes are compressed, with up to 30% of ribosomes in helical polysomes or collided disomes, some of which are bound to the stress effector GCN1. The native collision interface extends beyond the in vitro-characterized 40S and includes the L1 stalk and eEF2, possibly contributing to translocation inhibition. The accumulation of unresolved tRNA-bound 80S and 60S and aberrant 40S configurations identifies potentially limiting steps in collision responses. Our work provides a global view of the translation machinery in response to persistent collisions and a framework for quantitative analysis of translation dynamics in situ. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19222.map.gz emd_19222.map.gz | 6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19222-v30.xml emd-19222-v30.xml emd-19222.xml emd-19222.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19222.png emd_19222.png | 34.9 KB | ||
| Filedesc metadata |  emd-19222.cif.gz emd-19222.cif.gz | 3.9 KB | ||
| Others |  emd_19222_half_map_1.map.gz emd_19222_half_map_1.map.gz emd_19222_half_map_2.map.gz emd_19222_half_map_2.map.gz | 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19222 http://ftp.pdbj.org/pub/emdb/structures/EMD-19222 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19222 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19222 | HTTPS FTP |
-Validation report
| Summary document |  emd_19222_validation.pdf.gz emd_19222_validation.pdf.gz | 908.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19222_full_validation.pdf.gz emd_19222_full_validation.pdf.gz | 908.4 KB | Display | |
| Data in XML |  emd_19222_validation.xml.gz emd_19222_validation.xml.gz | 8.9 KB | Display | |
| Data in CIF |  emd_19222_validation.cif.gz emd_19222_validation.cif.gz | 10.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19222 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19222 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19222 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19222 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19222.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19222.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.34 Å | ||||||||||||||||||||||||||||||||||||
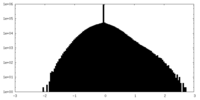
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_19222_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
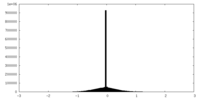
| Density Histograms |
-Half map: #2
| File | emd_19222_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
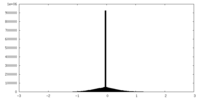
| Density Histograms |
- Sample components
Sample components
-Entire : in situ subtomogram average of MEF cell ribosome
| Entire | Name: in situ subtomogram average of MEF cell ribosome |
|---|---|
| Components |
|
-Supramolecule #1: in situ subtomogram average of MEF cell ribosome
| Supramolecule | Name: in situ subtomogram average of MEF cell ribosome / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: DMEM |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
| Details | MEF cells |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 2.3 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 63000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.5 µm / Nominal defocus min: 2.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 28.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number subtomograms used: 366 |
|---|---|
| Extraction | Number tomograms: 96 / Number images used: 1388 |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)