[English] 日本語
 Yorodumi
Yorodumi- EMDB-19129: A DNA Robotic Switch with Regulated Autonomous Display of Cytotox... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | A DNA Robotic Switch with Regulated Autonomous Display of Cytotoxic Ligand Nanopatterns | |||||||||||||||||||||
 Map data Map data | From cryoSPARC 3DFlex reconstruct job | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | DNA / DNA origami / Lgand nanopattern / pH sensitive switch / Solid tumor / Death receptor clustering | |||||||||||||||||||||
| Biological species | synthetic construct (others) /  Phage M13mp18 (virus) Phage M13mp18 (virus) | |||||||||||||||||||||
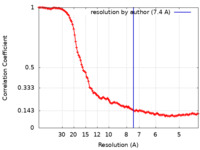
| Method | single particle reconstruction / cryo EM / Resolution: 7.4 Å | |||||||||||||||||||||
 Authors Authors | Wang Y / Berzina I / Hogberg B | |||||||||||||||||||||
| Funding support |  Sweden, European Union, Sweden, European Union,  Finland, 6 items Finland, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Nanotechnol / Year: 2024 Journal: Nat Nanotechnol / Year: 2024Title: A DNA robotic switch with regulated autonomous display of cytotoxic ligand nanopatterns. Authors: Yang Wang / Igor Baars / Ieva Berzina / Iris Rocamonde-Lago / Boxuan Shen / Yunshi Yang / Marco Lolaico / Janine Waldvogel / Ioanna Smyrlaki / Keying Zhu / Robert A Harris / Björn Högberg /   Abstract: The clustering of death receptors (DRs) at the membrane leads to apoptosis. With the goal of treating tumours, multivalent molecular tools that initiate this mechanism have been developed. However, ...The clustering of death receptors (DRs) at the membrane leads to apoptosis. With the goal of treating tumours, multivalent molecular tools that initiate this mechanism have been developed. However, DRs are also ubiquitously expressed in healthy tissue. Here we present a stimuli-responsive robotic switch nanodevice that can autonomously and selectively turn on the display of cytotoxic ligand patterns in tumour microenvironments. We demonstrate a switchable DNA origami that normally hides six ligands but displays them as a hexagonal pattern 10 nm in diameter once under higher acidity. This can effectively cluster DRs and trigger apoptosis of human breast cancer cells at pH 6.5 while remaining inert at pH 7.4. When administered to mice bearing human breast cancer xenografts, this nanodevice decreased tumour growth by up to 70%. The data demonstrate the feasibility and opportunities for developing ligand pattern switches as a path for targeted treatment. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19129.map.gz emd_19129.map.gz | 117.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19129-v30.xml emd-19129-v30.xml emd-19129.xml emd-19129.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19129_fsc.xml emd_19129_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_19129.png emd_19129.png | 62 KB | ||
| Filedesc metadata |  emd-19129.cif.gz emd-19129.cif.gz | 4.5 KB | ||
| Others |  emd_19129_half_map_1.map.gz emd_19129_half_map_1.map.gz emd_19129_half_map_2.map.gz emd_19129_half_map_2.map.gz | 14.5 MB 14.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19129 http://ftp.pdbj.org/pub/emdb/structures/EMD-19129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19129 | HTTPS FTP |
-Validation report
| Summary document |  emd_19129_validation.pdf.gz emd_19129_validation.pdf.gz | 657.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19129_full_validation.pdf.gz emd_19129_full_validation.pdf.gz | 656.7 KB | Display | |
| Data in XML |  emd_19129_validation.xml.gz emd_19129_validation.xml.gz | 18.4 KB | Display | |
| Data in CIF |  emd_19129_validation.cif.gz emd_19129_validation.cif.gz | 24.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19129 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19129 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19129 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19129 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19129.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19129.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | From cryoSPARC 3DFlex reconstruct job | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||
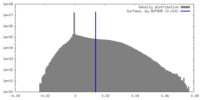
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: From cryoSPARC 3DFlex reconstruct job
| File | emd_19129_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | From cryoSPARC 3DFlex reconstruct job | ||||||||||||
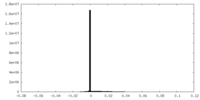
| Projections & Slices |
| ||||||||||||
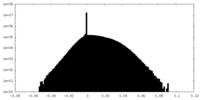
| Density Histograms |
-Half map: From cryoSPARC 3DFlex reconstruct job
| File | emd_19129_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | From cryoSPARC 3DFlex reconstruct job | ||||||||||||
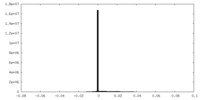
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DNA origami
| Entire | Name: DNA origami |
|---|---|
| Components |
|
-Supramolecule #1: DNA origami
| Supramolecule | Name: DNA origami / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Supramolecule #2: ssDNA staples
| Supramolecule | Name: ssDNA staples / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Supramolecule #3: ssDNA scaffold
| Supramolecule | Name: ssDNA scaffold / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Phage M13mp18 (virus) Phage M13mp18 (virus) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 5 mM MgCl2, 5 mM NaCl, 5 mM TRIS, and 1 mM EDTA |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 296 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Slit width: 10 eV |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 (6k x 4k) / #0 - Number grids imaged: 1 / #0 - Number real images: 6652 / #0 - Average exposure time: 4.4 sec. / #0 - Average electron dose: 40.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 (6k x 4k) / #1 - Number grids imaged: 1 / #1 - Number real images: 5195 / #1 - Average exposure time: 6.0 sec. / #1 - Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)