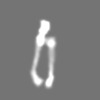[English] 日本語
 Yorodumi
Yorodumi- EMDB-1888: Thermus thermophilus V-type (A-type) ATPase at 16 Angstroms resolution -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1888 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Thermus thermophilus V-type (A-type) ATPase at 16 Angstroms resolution | |||||||||
 Map data Map data | Thermus thermophilus V-type (A-type) ATPase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Vacuolar type ATPase / V-ATPase / A-ATPase / Thermus thermophilus / ATP synthase / membrane protein | |||||||||
| Biological species |   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Lau WCY / Rubinstein JL | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2010 Journal: Proc Natl Acad Sci U S A / Year: 2010Title: Structure of intact Thermus thermophilus V-ATPase by cryo-EM reveals organization of the membrane-bound V(O) motor. Authors: Wilson C Y Lau / John L Rubinstein /  Abstract: The eubacterium Thermus thermophilus uses a macromolecular assembly closely related to eukaryotic V-ATPase to produce its supply of ATP. This simplified V-ATPase offers several advantages over ...The eubacterium Thermus thermophilus uses a macromolecular assembly closely related to eukaryotic V-ATPase to produce its supply of ATP. This simplified V-ATPase offers several advantages over eukaryotic V-ATPases for structural analysis and investigation of the mechanism of the enzyme. Here we report the structure of the complex at approximately 16 A resolution as determined by single particle electron cryomicroscopy (cryo-EM). The resolution of the map and our use of cryo-EM, rather than negative stain EM, reveals detailed information about the internal organization of the assembly. We could separate the map into segments corresponding to subunits A and B, the threefold pseudosymmetric C-subunit, a central rotor consisting of subunits D and F, the L-ring, the stator subcomplex consisting of subunits I, E, and G, and a micelle of bound detergent. The architecture of the V(O) region shows a remarkably small area of contact between the I-subunit and the ring of L-subunits and is consistent with a two half-channel model for proton translocation. The arrangement of structural elements in V(O) gives insight into the mechanism of torque generation from proton translocation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1888.map.gz emd_1888.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1888-v30.xml emd-1888-v30.xml emd-1888.xml emd-1888.xml | 8.2 KB 8.2 KB | Display Display |  EMDB header EMDB header |
| Images |  1888.jpg 1888.jpg | 91.3 KB | ||
| Others |  Supplementary_Map_Information.txt Supplementary_Map_Information.txt emd_1888_additional_1.map.gz emd_1888_additional_1.map.gz emd_1888_additional_10.map.gz emd_1888_additional_10.map.gz emd_1888_additional_11.map.gz emd_1888_additional_11.map.gz emd_1888_additional_2.map.gz emd_1888_additional_2.map.gz emd_1888_additional_3.map.gz emd_1888_additional_3.map.gz emd_1888_additional_4.map.gz emd_1888_additional_4.map.gz emd_1888_additional_5.map.gz emd_1888_additional_5.map.gz emd_1888_additional_6.map.gz emd_1888_additional_6.map.gz emd_1888_additional_7.map.gz emd_1888_additional_7.map.gz emd_1888_additional_8.map.gz emd_1888_additional_8.map.gz emd_1888_additional_9.map.gz emd_1888_additional_9.map.gz | 904 B 7.4 MB 7.4 MB 7.4 MB 7.4 MB 7.4 MB 7.4 MB 7.4 MB 7.4 MB 7.4 MB 7.4 MB 7.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1888 http://ftp.pdbj.org/pub/emdb/structures/EMD-1888 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1888 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1888 | HTTPS FTP |
-Validation report
| Summary document |  emd_1888_validation.pdf.gz emd_1888_validation.pdf.gz | 214.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1888_full_validation.pdf.gz emd_1888_full_validation.pdf.gz | 213.7 KB | Display | |
| Data in XML |  emd_1888_validation.xml.gz emd_1888_validation.xml.gz | 5.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1888 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1888 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1888 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1888 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1888.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1888.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Thermus thermophilus V-type (A-type) ATPase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
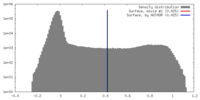
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Supplemental map: emd 1888 additional 1.map
+Supplemental map: emd 1888 additional 10.map
+Supplemental map: emd 1888 additional 11.map
+Supplemental map: emd 1888 additional 2.map
+Supplemental map: emd 1888 additional 3.map
+Supplemental map: emd 1888 additional 4.map
+Supplemental map: emd 1888 additional 5.map
+Supplemental map: emd 1888 additional 6.map
+Supplemental map: emd 1888 additional 7.map
+Supplemental map: emd 1888 additional 8.map
+Supplemental map: emd 1888 additional 9.map
+Others
- Sample components
Sample components
-Entire : V/A-ATPase from Thermus thermophilus solubilized with the deterge...
| Entire | Name: V/A-ATPase from Thermus thermophilus solubilized with the detergent dodecyl maltoside. |
|---|---|
| Components |
|
-Supramolecule #1000: V/A-ATPase from Thermus thermophilus solubilized with the deterge...
| Supramolecule | Name: V/A-ATPase from Thermus thermophilus solubilized with the detergent dodecyl maltoside. type: sample / ID: 1000 / Oligomeric state: Hetero 26mer / Number unique components: 9 |
|---|---|
| Molecular weight | Theoretical: 600 KDa |
-Supramolecule #1: V-ATPase
| Supramolecule | Name: V-ATPase / type: organelle_or_cellular_component / ID: 1 / Name.synonym: ATPase / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) / Location in cell: Plasma membrane Thermus thermophilus (bacteria) / Location in cell: Plasma membrane |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 50mM Tris-HCl 150mM NaCL 5mM MgCl2 O.02% DDM |
| Grid | Details: Quantifoil 2/2 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK III / Details: Vitrification instrument: Vitrobot Mark III / Method: Blot approx. 20 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7.0 µm / Average electron dose: 25 e/Å2 / Details: Images averaged 2x2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Gatan 626 / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Particle images were manually selected with Ximdisp. |
|---|---|
| CTF correction | Details: MRC |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 16.0 Å / Resolution method: OTHER / Software - Name: Rotan, Frealign / Number images used: 19825 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)