[English] 日本語
 Yorodumi
Yorodumi- EMDB-18820: Structure of avian H5N1 influenza A polymerase dimer in complex w... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of avian H5N1 influenza A polymerase dimer in complex with human ANP32B (Encapsidase focused). | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Influenza virus / replication platform / H5N1 / influenza adaptive mutations / host adaptation / viral polymerase / cryo-EM / HPAI. / VIRAL PROTEIN | |||||||||||||||||||||
| Biological species |   Influenza A virus Influenza A virus | |||||||||||||||||||||
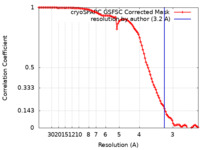
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||||||||
 Authors Authors | Carrique L / Staller E / Keown JR / Fan H / Fodor E / Grimes JM | |||||||||||||||||||||
| Funding support |  United Kingdom, 6 items United Kingdom, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structures of H5N1 influenza polymerase with ANP32B reveal mechanisms of genome replication and host adaptation. Authors: Ecco Staller / Loïc Carrique / Olivia C Swann / Haitian Fan / Jeremy R Keown / Carol M Sheppard / Wendy S Barclay / Jonathan M Grimes / Ervin Fodor /   Abstract: Avian influenza A viruses (IAVs) pose a public health threat, as they are capable of triggering pandemics by crossing species barriers. Replication of avian IAVs in mammalian cells is hindered by ...Avian influenza A viruses (IAVs) pose a public health threat, as they are capable of triggering pandemics by crossing species barriers. Replication of avian IAVs in mammalian cells is hindered by species-specific variation in acidic nuclear phosphoprotein 32 (ANP32) proteins, which are essential for viral RNA genome replication. Adaptive mutations enable the IAV RNA polymerase (FluPolA) to surmount this barrier. Here, we present cryo-electron microscopy structures of monomeric and dimeric avian H5N1 FluPolA with human ANP32B. ANP32B interacts with the PA subunit of FluPolA in the monomeric form, at the site used for its docking onto the C-terminal domain of host RNA polymerase II during viral transcription. ANP32B acts as a chaperone, guiding FluPolA towards a ribonucleoprotein-associated FluPolA to form an asymmetric dimer-the replication platform for the viral genome. These findings offer insights into the molecular mechanisms governing IAV genome replication, while enhancing our understanding of the molecular processes underpinning mammalian adaptations in avian-origin FluPolA. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18820.map.gz emd_18820.map.gz | 97.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18820-v30.xml emd-18820-v30.xml emd-18820.xml emd-18820.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18820_fsc.xml emd_18820_fsc.xml | 9.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_18820.png emd_18820.png | 49.3 KB | ||
| Masks |  emd_18820_msk_1.map emd_18820_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18820.cif.gz emd-18820.cif.gz | 4.8 KB | ||
| Others |  emd_18820_half_map_1.map.gz emd_18820_half_map_1.map.gz emd_18820_half_map_2.map.gz emd_18820_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18820 http://ftp.pdbj.org/pub/emdb/structures/EMD-18820 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18820 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18820 | HTTPS FTP |
-Validation report
| Summary document |  emd_18820_validation.pdf.gz emd_18820_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18820_full_validation.pdf.gz emd_18820_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_18820_validation.xml.gz emd_18820_validation.xml.gz | 18.3 KB | Display | |
| Data in CIF |  emd_18820_validation.cif.gz emd_18820_validation.cif.gz | 23.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18820 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18820 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18820 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18820 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18820.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18820.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2427 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18820_msk_1.map emd_18820_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18820_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18820_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Avian H5N1 influenza A replication platform in complex with human...
| Entire | Name: Avian H5N1 influenza A replication platform in complex with human ANP32B. |
|---|---|
| Components |
|
-Supramolecule #1: Avian H5N1 influenza A replication platform in complex with human...
| Supramolecule | Name: Avian H5N1 influenza A replication platform in complex with human ANP32B. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:   Influenza A virus / Strain: A/turkey/Turkey/1/2005(H5N1) Influenza A virus / Strain: A/turkey/Turkey/1/2005(H5N1) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 24.0 µm / Nominal defocus min: 5.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)