[English] 日本語
 Yorodumi
Yorodumi- EMDB-18812: The structure of the human 80S ribosome at 1.9 angstrom resolutio... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of the human 80S ribosome at 1.9 angstrom resolution reveals the molecular role of chemical modifications and ions in RNA - Focused refinement of the of the 60S subunit | |||||||||||||||
 Map data Map data | Focused refinement of the 60S ribosome subunit using the multibody job in relion. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | ribosome / cryo-electron microscopie / post-transcriptional modifications | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.9 Å | |||||||||||||||
 Authors Authors | Holvec S / Barchet C / Frechin L / Hazemann I / von Loeffelholz O / Klaholz BP | |||||||||||||||
| Funding support |  France, 4 items France, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: The structure of the human 80S ribosome at 1.9 Å resolution reveals the molecular role of chemical modifications and ions in RNA. Authors: Samuel Holvec / Charles Barchet / Antony Lechner / Léo Fréchin / S Nimali T De Silva / Isabelle Hazemann / Philippe Wolff / Ottilie von Loeffelholz / Bruno P Klaholz /  Abstract: The ribosomal RNA of the human protein synthesis machinery comprises numerous chemical modifications that are introduced during ribosome biogenesis. Here we present the 1.9 Å resolution cryo ...The ribosomal RNA of the human protein synthesis machinery comprises numerous chemical modifications that are introduced during ribosome biogenesis. Here we present the 1.9 Å resolution cryo electron microscopy structure of the 80S human ribosome resolving numerous new ribosomal RNA modifications and functionally important ions such as Zn, K and Mg, including their associated individual water molecules. The 2'-O-methylation, pseudo-uridine and base modifications were confirmed by mass spectrometry, resulting in a complete investigation of the >230 sites, many of which could not be addressed previously. They choreograph key interactions within the RNA and at the interface with proteins, including at the ribosomal subunit interfaces of the fully assembled 80S ribosome. Uridine isomerization turns out to be a key mechanism for U-A base pair stabilization in RNA in general. The structural environment of chemical modifications and ions is primordial for the RNA architecture of the mature human ribosome, hence providing a structural framework to address their role in healthy states and in human diseases. #1:  Journal: Biorxiv / Year: 2023 Journal: Biorxiv / Year: 2023Title: Structure of the human 80S ribosome at 1.9 angstrom resolution - the molecular role of chemical modifications and ions in RNA Authors: Holvec S / Barchet C / Lechner A / Frechin L / Silva SNTD / Hazemann I / Wolff P / Loeffelholz OV / Klaholz BP | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18812.map.gz emd_18812.map.gz | 46.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18812-v30.xml emd-18812-v30.xml emd-18812.xml emd-18812.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18812.png emd_18812.png | 55.4 KB | ||
| Filedesc metadata |  emd-18812.cif.gz emd-18812.cif.gz | 4.4 KB | ||
| Others |  emd_18812_half_map_1.map.gz emd_18812_half_map_1.map.gz emd_18812_half_map_2.map.gz emd_18812_half_map_2.map.gz | 279.9 MB 280.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18812 http://ftp.pdbj.org/pub/emdb/structures/EMD-18812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18812 | HTTPS FTP |
-Validation report
| Summary document |  emd_18812_validation.pdf.gz emd_18812_validation.pdf.gz | 794.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18812_full_validation.pdf.gz emd_18812_full_validation.pdf.gz | 793.8 KB | Display | |
| Data in XML |  emd_18812_validation.xml.gz emd_18812_validation.xml.gz | 18.6 KB | Display | |
| Data in CIF |  emd_18812_validation.cif.gz emd_18812_validation.cif.gz | 22.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18812 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18812 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18812 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18812 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18812.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18812.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused refinement of the 60S ribosome subunit using the multibody job in relion. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||
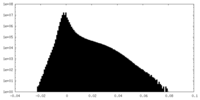
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Second half-map of the 60S subunit map.
| File | emd_18812_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second half-map of the 60S subunit map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
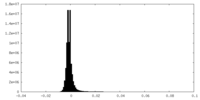
| Density Histograms |
- Sample components
Sample components
-Entire : Human 60S ribosomal subunit
| Entire | Name: Human 60S ribosomal subunit |
|---|---|
| Components |
|
-Supramolecule #1: Human 60S ribosomal subunit
| Supramolecule | Name: Human 60S ribosomal subunit / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.6 |
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV |
| Details | Purified from HeLa cells. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 6528 / Average exposure time: 3.58 sec. / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.1 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 1.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 382016 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)