[English] 日本語
 Yorodumi
Yorodumi- EMDB-18774: Cryo-EM map of calcium-bound mTMEM16A(ac)-L647V/I733V chloride ch... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of calcium-bound mTMEM16A(ac)-L647V/I733V chloride channel at 3.29 A resolution | |||||||||
 Map data Map data | Cryo-EM map of calcium-bound mTMEM16A(ac)-L647V/I733V chloride channel at 3.29 A resolution | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | calcium-activated chloride channel / anoctamin-1 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationglial cell projection elongation / trachea development / iodide transmembrane transporter activity / iodide transport / intracellularly calcium-gated chloride channel activity / mucus secretion / cellular response to peptide / voltage-gated chloride channel activity / Stimuli-sensing channels / chloride transport ...glial cell projection elongation / trachea development / iodide transmembrane transporter activity / iodide transport / intracellularly calcium-gated chloride channel activity / mucus secretion / cellular response to peptide / voltage-gated chloride channel activity / Stimuli-sensing channels / chloride transport / chloride channel activity / positive regulation of insulin secretion involved in cellular response to glucose stimulus / detection of temperature stimulus involved in sensory perception of pain / chloride channel complex / chloride transmembrane transport / regulation of membrane potential / establishment of localization in cell / cell projection / presynaptic membrane / phospholipase C-activating G protein-coupled receptor signaling pathway / cellular response to heat / apical plasma membrane / external side of plasma membrane / signaling receptor binding / glutamatergic synapse / protein homodimerization activity / identical protein binding / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.29 Å | |||||||||
 Authors Authors | Lam AKM / Dutzler R | |||||||||
| Funding support | European Union,  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2023 Journal: EMBO J / Year: 2023Title: Mechanistic basis of ligand efficacy in the calcium-activated chloride channel TMEM16A. Authors: Andy Km Lam / Raimund Dutzler /  Abstract: Agonist binding in ligand-gated ion channels is coupled to structural rearrangements around the binding site, followed by the opening of the channel pore. In this process, agonist efficacy describes ...Agonist binding in ligand-gated ion channels is coupled to structural rearrangements around the binding site, followed by the opening of the channel pore. In this process, agonist efficacy describes the equilibrium between open and closed conformations in a fully ligand-bound state. Calcium-activated chloride channels in the TMEM16 family are important sensors of intracellular calcium signals and are targets for pharmacological modulators, yet a mechanistic understanding of agonist efficacy has remained elusive. Using a combination of cryo-electron microscopy, electrophysiology, and autocorrelation analysis, we now show that agonist efficacy in the ligand-gated channel TMEM16A is dictated by the conformation of the pore-lining helix α6 around the Ca -binding site. The closure of the binding site, which involves the formation of a π-helix below a hinge region in α6, appears to be coupled to the opening of the inner pore gate, thereby governing the channel's open probability and conductance. Our results provide a mechanism for agonist binding and efficacy and a structural basis for the design of potentiators and partial agonists in the TMEM16 family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18774.map.gz emd_18774.map.gz | 12.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18774-v30.xml emd-18774-v30.xml emd-18774.xml emd-18774.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18774.png emd_18774.png | 81.2 KB | ||
| Masks |  emd_18774_msk_1.map emd_18774_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18774.cif.gz emd-18774.cif.gz | 6 KB | ||
| Others |  emd_18774_half_map_1.map.gz emd_18774_half_map_1.map.gz emd_18774_half_map_2.map.gz emd_18774_half_map_2.map.gz | 159.9 MB 159.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18774 http://ftp.pdbj.org/pub/emdb/structures/EMD-18774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18774 | HTTPS FTP |
-Related structure data
| Related structure data |  8qzcMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18774.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18774.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of calcium-bound mTMEM16A(ac)-L647V/I733V chloride channel at 3.29 A resolution | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.659 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18774_msk_1.map emd_18774_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
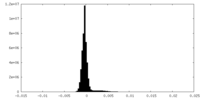
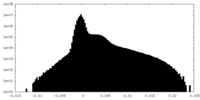
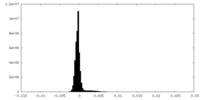
| Density Histograms |
-Half map: Half-map 2 of calcium-bound mTMEM16A(ac)-L647V/I733V chloride channel
| File | emd_18774_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 of calcium-bound mTMEM16A(ac)-L647V/I733V chloride channel | ||||||||||||
| Projections & Slices |
| ||||||||||||
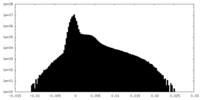
| Density Histograms |
-Half map: Half-map 1 of calcium-bound mTMEM16A(ac)-L647V/I733V chloride channel
| File | emd_18774_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 of calcium-bound mTMEM16A(ac)-L647V/I733V chloride channel | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mouse TMEM16A ac splice variant with introduced mutations L647V a...
| Entire | Name: mouse TMEM16A ac splice variant with introduced mutations L647V and I733V in complex with calcium |
|---|---|
| Components |
|
-Supramolecule #1: mouse TMEM16A ac splice variant with introduced mutations L647V a...
| Supramolecule | Name: mouse TMEM16A ac splice variant with introduced mutations L647V and I733V in complex with calcium type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Anoctamin-1
| Macromolecule | Name: Anoctamin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 111.030945 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRVPEKYSTL PAEDRSVHIV NICAIEDLGY LPSEGTLLNS LSVDPDAECK YGLYFRDGKR KVDYILVYHH KRASGSRTLA RRGLQNDMV LGTRSVRQDQ PLPGKGSPVD AGSPEVPMDY HEDDKRFRRE EYEGNLLEAG LELENDEDTK IHGVGFVKIH A PWHVLCRE ...String: MRVPEKYSTL PAEDRSVHIV NICAIEDLGY LPSEGTLLNS LSVDPDAECK YGLYFRDGKR KVDYILVYHH KRASGSRTLA RRGLQNDMV LGTRSVRQDQ PLPGKGSPVD AGSPEVPMDY HEDDKRFRRE EYEGNLLEAG LELENDEDTK IHGVGFVKIH A PWHVLCRE AEFLKLKMPT KKVYHISETR GLLKTINSVL QKITDPIQPK VAEHRPQTTK RLSYPFSREK QHLFDLTDRD SF FDSKTRS TIVYEILKRT TCTKAKYSMG ITSLLANGVY SAAYPLHDGD YEGDNVEFND RKLLYEEWAS YGVFYKYQPI DLV RKYFGE KVGLYFAWLG AYTQMLIPAS IVGVIVFLYG CATVDENIPS MEMCDQRYNI TMCPLCDKTC SYWKMSSACA TARA SHLFD NPATVFFSVF MALWAATFME HWKRKQMRLN YRWDLTGFEE EEEAVKDHPR AEYEARVLEK SLRKESRNKE TDKVK LTWR DRFPAYFTNL VSIIFMIAVT FAIVLGVIIY RISTAAALAM NSSPSVRSNI RVTVTATAVI INLVVIILLD EVYGCI ARW LTKIEVPKTE KSFEERLTFK AFLLKFVNSY TPIFYVAFFK GRFVGRPGDY VYIFRSFRME ECAPGGCLME LCIQLSI IM LGKQVIQNNL FEIGIPKMKK FIRYLKLRRQ SPSDREEYVK RKQRYEVDFN LEPFAGLTPE YMEMIIQFGF VTLFVASF P LAPLFALLNN IVEIRLDAKK FVTELRRPVA IRAKDIGIWY NILRGVGKLA VIINAFVISF TSDFIPRLVY LYMYSQNGT MHGFVNHTLS SFNVSDFQNG TAPNDPLDLG YEVQICRYKD YREPPWSEHK YDISKDFWAV LAARLAFVIV FQNLVMFMSD FVDWVIPDI PKDISQQIHK EKVLMVELFM REEQGKQQLL DTWMEKEKPR DVPCNNHSPT THPEAGDGSP VPSYEYHGDA L UniProtKB: Anoctamin-1 |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 62.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.29 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 144934 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



 Z
Z Y
Y X
X