[English] 日本語
 Yorodumi
Yorodumi- EMDB-18327: CryoEM structure of a S. Cerevisiae Ski2387 complex in an interme... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of a S. Cerevisiae Ski2387 complex in an intermediate state bound to RNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Helicase / RNA binding / RNA degradation / HYDROLASE | |||||||||
| Biological species |  | |||||||||
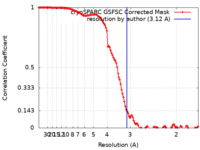
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | |||||||||
 Authors Authors | Keidel A / Koegel A / Reichelt P / Kowalinski E / Schaefer IB / Conti E | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly. Authors: Achim Keidel / Alexander Kögel / Peter Reichelt / Eva Kowalinski / Ingmar B Schäfer / Elena Conti /   Abstract: The Ski2-Ski3-Ski8 (Ski238) helicase complex directs cytoplasmic mRNAs toward the nucleolytic exosome complex for degradation. In yeast, the interaction between Ski238 and exosome requires the ...The Ski2-Ski3-Ski8 (Ski238) helicase complex directs cytoplasmic mRNAs toward the nucleolytic exosome complex for degradation. In yeast, the interaction between Ski238 and exosome requires the adaptor protein Ski7. We determined different cryo-EM structures of the Ski238 complex depicting the transition from a rigid autoinhibited closed conformation to a flexible active open conformation in which the Ski2 helicase module has detached from the rest of Ski238. The open conformation favors the interaction of the Ski3 subunit with exosome-bound Ski7, leading to the recruitment of the exosome. In the Ski238-Ski7-exosome holocomplex, the Ski2 helicase module binds the exosome cap, enabling the RNA to traverse from the helicase through the internal exosome channel to the Rrp44 exoribonuclease. Our study pinpoints how conformational changes within the Ski238 complex regulate exosome recruitment for RNA degradation. We also reveal the remarkable conservation of helicase-exosome RNA channeling mechanisms throughout eukaryotic nuclear and cytoplasmic exosome complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18327.map.gz emd_18327.map.gz | 157.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18327-v30.xml emd-18327-v30.xml emd-18327.xml emd-18327.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18327_fsc.xml emd_18327_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_18327.png emd_18327.png | 39.8 KB | ||
| Masks |  emd_18327_msk_1.map emd_18327_msk_1.map | 166.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18327.cif.gz emd-18327.cif.gz | 4.2 KB | ||
| Others |  emd_18327_additional_1.map.gz emd_18327_additional_1.map.gz emd_18327_half_map_1.map.gz emd_18327_half_map_1.map.gz emd_18327_half_map_2.map.gz emd_18327_half_map_2.map.gz | 148 MB 154.3 MB 154.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18327 http://ftp.pdbj.org/pub/emdb/structures/EMD-18327 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18327 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18327 | HTTPS FTP |
-Validation report
| Summary document |  emd_18327_validation.pdf.gz emd_18327_validation.pdf.gz | 939.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18327_full_validation.pdf.gz emd_18327_full_validation.pdf.gz | 939.2 KB | Display | |
| Data in XML |  emd_18327_validation.xml.gz emd_18327_validation.xml.gz | 19.6 KB | Display | |
| Data in CIF |  emd_18327_validation.cif.gz emd_18327_validation.cif.gz | 25.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18327 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18327 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18327 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18327 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18327.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18327.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8512 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18327_msk_1.map emd_18327_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: sharpened with deepEMhancer software (tightTarget)
| File | emd_18327_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened with deepEMhancer software (tightTarget) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18327_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18327_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ski2delarch387 PolyU RNA
| Entire | Name: Ski2delarch387 PolyU RNA |
|---|---|
| Components |
|
-Supramolecule #1: Ski2delarch387 PolyU RNA
| Supramolecule | Name: Ski2delarch387 PolyU RNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 9416 / Average electron dose: 67.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)