[English] 日本語
 Yorodumi
Yorodumi- EMDB-18010: Charging of vitreous samples in cryogenic electron microscopy mit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Charging of vitreous samples in cryogenic electron microscopy mitigated by graphene - BfrB - Dataset 4 - Quantifoil 300 mesh R1.2/1.3 with Graphene - Large Beam | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | IRON STORAGE / FERROXIDASE / BACTERIAL FERRITIN / OCTAHEDRAL SYMMETRY. / METAL TRANSPORT / METAL BINDING PROTEIN | ||||||||||||
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.9 Å | ||||||||||||
 Authors Authors | van schayck JP / Zhang Y / Ravelli RBG | ||||||||||||
| Funding support |  Netherlands, European Union, 3 items Netherlands, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: ACS Nano / Year: 2023 Journal: ACS Nano / Year: 2023Title: Charging of Vitreous Samples in Cryogenic Electron Microscopy Mitigated by Graphene. Authors: Yue Zhang / J Paul van Schayck / Adrián Pedrazo-Tardajos / Nathalie Claes / Willem E M Noteborn / Peng-Han Lu / Hans Duimel / Rafal E Dunin-Borkowski / Sara Bals / Peter J Peters / Raimond B G Ravelli /    Abstract: Cryogenic electron microscopy can provide high-resolution reconstructions of macromolecules embedded in a thin layer of ice from which atomic models can be built . However, the interaction between ...Cryogenic electron microscopy can provide high-resolution reconstructions of macromolecules embedded in a thin layer of ice from which atomic models can be built . However, the interaction between the ionizing electron beam and the sample results in beam-induced motion and image distortion, which limit the attainable resolutions. Sample charging is one contributing factor of beam-induced motions and image distortions, which is normally alleviated by including part of the supporting conducting film within the beam-exposed region. However, routine data collection schemes avoid strategies whereby the beam is not in contact with the supporting film, whose rationale is not fully understood. Here we characterize electrostatic charging of vitreous samples, both in imaging and in diffraction mode. We mitigate sample charging by depositing a single layer of conductive graphene on top of regular EM grids. We obtained high-resolution single-particle analysis (SPA) reconstructions at 2 Å when the electron beam only irradiates the middle of the hole on graphene-coated grids, using data collection schemes that previously failed to produce sub 3 Å reconstructions without the graphene layer. We also observe that the SPA data obtained with the graphene-coated grids exhibit a higher factor and reduced particle movement compared to data obtained without the graphene layer. This mitigation of charging could have broad implications for various EM techniques, including SPA and cryotomography, and for the study of radiation damage and the development of future sample carriers. Furthermore, it may facilitate the exploration of more dose-efficient, scanning transmission EM based SPA techniques. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18010.map.gz emd_18010.map.gz | 121.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18010-v30.xml emd-18010-v30.xml emd-18010.xml emd-18010.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18010_fsc.xml emd_18010_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_18010.png emd_18010.png | 855.6 KB | ||
| Others |  emd_18010_half_map_1.map.gz emd_18010_half_map_1.map.gz emd_18010_half_map_2.map.gz emd_18010_half_map_2.map.gz | 97.7 MB 97.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18010 http://ftp.pdbj.org/pub/emdb/structures/EMD-18010 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18010 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18010 | HTTPS FTP |
-Validation report
| Summary document |  emd_18010_validation.pdf.gz emd_18010_validation.pdf.gz | 840.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18010_full_validation.pdf.gz emd_18010_full_validation.pdf.gz | 839.8 KB | Display | |
| Data in XML |  emd_18010_validation.xml.gz emd_18010_validation.xml.gz | 18.5 KB | Display | |
| Data in CIF |  emd_18010_validation.cif.gz emd_18010_validation.cif.gz | 24 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18010 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18010 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18010 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18010 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18010.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18010.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.834 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_18010_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18010_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MYCOBACTERIUM TUBERCULOSIS FERRITIN
| Entire | Name: MYCOBACTERIUM TUBERCULOSIS FERRITIN |
|---|---|
| Components |
|
-Supramolecule #1: MYCOBACTERIUM TUBERCULOSIS FERRITIN
| Supramolecule | Name: MYCOBACTERIUM TUBERCULOSIS FERRITIN / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: MYCOBACTERIUM TUBERCULOSIS FERRITIN |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
-Macromolecule #1: Ferritin BfrB
| Macromolecule | Name: Ferritin BfrB / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: ferroxidase |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MTEYEGPKTK FHALMQEQI H NEFTAAQQ YV AIAVYFD SED LPQLAK HFYS QAVEE RNHAM MLVQ HLLDRD LRV EIPGVDT VR NQFDRPRE A LALALDQER TVTDQVGRLT AVARDEGDF L GEQFMQWF LQ EQIEEVA LMA TLVRVA DRAG ANLFE ...String: MTEYEGPKTK FHALMQEQI H NEFTAAQQ YV AIAVYFD SED LPQLAK HFYS QAVEE RNHAM MLVQ HLLDRD LRV EIPGVDT VR NQFDRPRE A LALALDQER TVTDQVGRLT AVARDEGDF L GEQFMQWF LQ EQIEEVA LMA TLVRVA DRAG ANLFE LENFV AREV DVAPAA SGA PHAAGGR L |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 6000 pixel / Digitization - Dimensions - Height: 4000 pixel / Number grids imaged: 1 / Number real images: 1808 / Average exposure time: 1.7 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


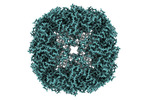



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































