[English] 日本語
 Yorodumi
Yorodumi- EMDB-17762: Cryo-EM structure of active Phthaloyl-CoA decarboxylase (Pcd) com... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of active Phthaloyl-CoA decarboxylase (Pcd) complex with prFMN bound | |||||||||
 Map data Map data | PCD holo-complex with prFMN co-factor bound | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | prFMN / plastic degradation / Phthalates / anaerobic / light sensitive / Co-enzyme A / decarboxylase / FLAVOPROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationferulate metabolic process / cinnamic acid catabolic process / carboxy-lyase activity / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Thauera chlorobenzoica (bacteria) Thauera chlorobenzoica (bacteria) | |||||||||
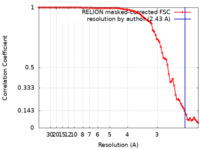
| Method | single particle reconstruction / cryo EM / Resolution: 2.43 Å | |||||||||
 Authors Authors | Kayastha K / Ermler U | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Active Pcd complex with prFMN bound Authors: Kayastha K / Ermler U | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17762.map.gz emd_17762.map.gz | 40.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17762-v30.xml emd-17762-v30.xml emd-17762.xml emd-17762.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17762_fsc.xml emd_17762_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_17762.png emd_17762.png | 153.1 KB | ||
| Filedesc metadata |  emd-17762.cif.gz emd-17762.cif.gz | 6.1 KB | ||
| Others |  emd_17762_half_map_1.map.gz emd_17762_half_map_1.map.gz emd_17762_half_map_2.map.gz emd_17762_half_map_2.map.gz | 40.5 MB 40.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17762 http://ftp.pdbj.org/pub/emdb/structures/EMD-17762 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17762 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17762 | HTTPS FTP |
-Validation report
| Summary document |  emd_17762_validation.pdf.gz emd_17762_validation.pdf.gz | 698.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17762_full_validation.pdf.gz emd_17762_full_validation.pdf.gz | 698.3 KB | Display | |
| Data in XML |  emd_17762_validation.xml.gz emd_17762_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_17762_validation.cif.gz emd_17762_validation.cif.gz | 20.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17762 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17762 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17762 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17762 | HTTPS FTP |
-Related structure data
| Related structure data |  8pmkMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17762.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17762.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PCD holo-complex with prFMN co-factor bound | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.116 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_17762_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
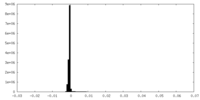
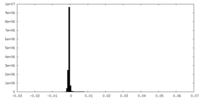
| Density Histograms |
-Half map: #1
| File | emd_17762_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
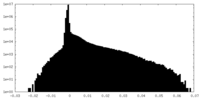
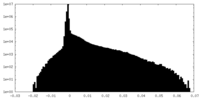
| Density Histograms |
- Sample components
Sample components
-Entire : Active Phthaloyl-CoA decarboxylase with prFMN co-factor bound
| Entire | Name: Active Phthaloyl-CoA decarboxylase with prFMN co-factor bound |
|---|---|
| Components |
|
-Supramolecule #1: Active Phthaloyl-CoA decarboxylase with prFMN co-factor bound
| Supramolecule | Name: Active Phthaloyl-CoA decarboxylase with prFMN co-factor bound type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Thauera chlorobenzoica (bacteria) Thauera chlorobenzoica (bacteria) |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: Phthaloyl-CoA decarboxylase
| Macromolecule | Name: Phthaloyl-CoA decarboxylase / type: protein_or_peptide / ID: 1 / Details: prFMN / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thauera chlorobenzoica (bacteria) Thauera chlorobenzoica (bacteria) |
| Molecular weight | Theoretical: 58.985941 KDa |
| Recombinant expression | Organism:  Thauera chlorobenzoica (bacteria) Thauera chlorobenzoica (bacteria) |
| Sequence | String: MNDLATKGIS EAAERVGEKD LRAALEWFRS KGYLVETNKE VNPDLEITGL QKIFDGSLPM LFNNVKDMPH ARAITNLFGD IRVVEELFG WENSLDRVKK VARAIDHPLK PVIIGQDEAP VQEEVLTTDL DVNKWLTAIR HTPLETEMTI GSGISCVVGP Y FDGGSHIG ...String: MNDLATKGIS EAAERVGEKD LRAALEWFRS KGYLVETNKE VNPDLEITGL QKIFDGSLPM LFNNVKDMPH ARAITNLFGD IRVVEELFG WENSLDRVKK VARAIDHPLK PVIIGQDEAP VQEEVLTTDL DVNKWLTAIR HTPLETEMTI GSGISCVVGP Y FDGGSHIG YNRMNFRWGN VGTFQISPGS HMWQVMTEHY KDDEPIPLTM CFGVPPSCTY VAGAGFDYAI LPKGCDEIGI AG AIQGSPV RLVKCRTIDA YTLADAEYVL EGYLHPRDKR YETAESEAAD IQGRFHFHPE WAGYMGKAYK APTFHVTAIT MRR RESKPI IFPLGVHTAD DANIDTSVRE SAIFALCERL QPGIVQNVHI PYCMTDWGGC IIQVKKRNQI EEGWQRNFLA AILA CSQGM RLAIAVSEDV DIYSMDDIMW CLTTRVNPQT DILNPLPGGR GQTFMPAERM TSGDKQWTAS NTQFEGGMGI DATVP YGYE SDFHRPVYGV DLVKPENFFD AKDIDKMKSR MAGWVLSLAR TGR UniProtKB: Phthaloyl-CoA decarboxylase |
-Macromolecule #2: hydroxylated prenyl-FMN
| Macromolecule | Name: hydroxylated prenyl-FMN / type: ligand / ID: 2 / Number of copies: 6 / Formula: BYN |
|---|---|
| Molecular weight | Theoretical: 542.476 Da |
| Chemical component information |  ChemComp-BYN: |
-Macromolecule #3: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 3 / Number of copies: 18 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #4: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 135 sec. / Pretreatment - Pressure: 38.0 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 30 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 9974 / Average exposure time: 2.82 sec. / Average electron dose: 50.76 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8pmk: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)