[English] 日本語
 Yorodumi
Yorodumi- EMDB-17749: The ERAD misfolded glycoprotein checkpoint complex from Chaetomiu... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The ERAD misfolded glycoprotein checkpoint complex from Chaetomium thermophilum (EDEM:PDI heterodimer). | ||||||||||||
 Map data Map data | Cryosparc class, 150,000 particles, iteration 4 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | mannosidase / disulfide isomerase / glycoprotein degradation / erad / misfolding / OXIDOREDUCTASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationendoplasmic reticulum mannose trimming / mannosyl-oligosaccharide 1,2-alpha-mannosidase activity / protein disulfide-isomerase / endoplasmic reticulum quality control compartment / Hydrolases; Glycosylases; Glycosidases, i.e. enzymes that hydrolyse O- and S-glycosyl compounds / protein disulfide isomerase activity / protein glycosylation / ERAD pathway / bioluminescence / generation of precursor metabolites and energy ...endoplasmic reticulum mannose trimming / mannosyl-oligosaccharide 1,2-alpha-mannosidase activity / protein disulfide-isomerase / endoplasmic reticulum quality control compartment / Hydrolases; Glycosylases; Glycosidases, i.e. enzymes that hydrolyse O- and S-glycosyl compounds / protein disulfide isomerase activity / protein glycosylation / ERAD pathway / bioluminescence / generation of precursor metabolites and energy / carbohydrate metabolic process / endoplasmic reticulum lumen / calcium ion binding / membrane Similarity search - Function | ||||||||||||
| Biological species |  Thermochaetoides thermophila (fungus) Thermochaetoides thermophila (fungus) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | ||||||||||||
 Authors Authors | Roversi P / Hitchman CJ / Lia A / Bayo Y | ||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Italy, 3 items Italy, 3 items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: The ERAD misfolded glycoprotein checkpoint complex from Chaetomium thermophilum (EDEM:PDI heterodimer). Authors: Roversi P / Hitchman CJ / Lia A / Bayo Y | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17749.map.gz emd_17749.map.gz | 51.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17749-v30.xml emd-17749-v30.xml emd-17749.xml emd-17749.xml | 24.8 KB 24.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17749.png emd_17749.png | 115.6 KB | ||
| Filedesc metadata |  emd-17749.cif.gz emd-17749.cif.gz | 8.2 KB | ||
| Others |  emd_17749_half_map_1.map.gz emd_17749_half_map_1.map.gz emd_17749_half_map_2.map.gz emd_17749_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17749 http://ftp.pdbj.org/pub/emdb/structures/EMD-17749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17749 | HTTPS FTP |
-Validation report
| Summary document |  emd_17749_validation.pdf.gz emd_17749_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17749_full_validation.pdf.gz emd_17749_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_17749_validation.xml.gz emd_17749_validation.xml.gz | 13.3 KB | Display | |
| Data in CIF |  emd_17749_validation.cif.gz emd_17749_validation.cif.gz | 15.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17749 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17749 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17749 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17749 | HTTPS FTP |
-Related structure data
| Related structure data |  8pkoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17749.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17749.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc class, 150,000 particles, iteration 4 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
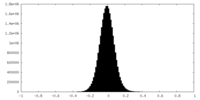
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryosparc class, 150,000 particles, iteration 3
| File | emd_17749_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc class, 150,000 particles, iteration 3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
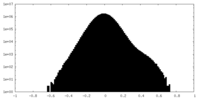
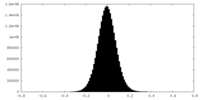
| Density Histograms |
-Half map: Cryosparc class, 150,000 particles, iteration 3
| File | emd_17749_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc class, 150,000 particles, iteration 3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Complex of Chaetomium thermophilum Endoplasmic reticulum degradat...
+Supramolecule #1: Complex of Chaetomium thermophilum Endoplasmic reticulum degradat...
+Supramolecule #2: Chaetomium thermophilum Endoplasmic reticulum degradation enhanci...
+Supramolecule #3: Endoplasmic reticulum degradation enhancing protein disulfide iso...
+Macromolecule #1: Green fluorescent protein,alpha-1,2-Mannosidase
+Macromolecule #2: Protein disulfide-isomerase
+Macromolecule #5: CALCIUM ION
+Macromolecule #6: THIOSULFATE
+Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #8: alpha-D-mannopyranose
+Macromolecule #9: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
Details: Micro SEC buffer: 150 mM NaCl, 20 mM MES pH 7.0, 1 mM CaCl2 | ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 180 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.2 kPa Details: The Quantifoil Au300 R1.2/1.3 Holey Carbon grids were glow discharged using the EMS GloQube. For grids to be coated in GO, the Graphene Oxide setting was used (0.2m Bar, 40 mA for 180 ...Details: The Quantifoil Au300 R1.2/1.3 Holey Carbon grids were glow discharged using the EMS GloQube. For grids to be coated in GO, the Graphene Oxide setting was used (0.2m Bar, 40 mA for 180 seconds). For GO coating, graphene oxide solution (Sigma) at 0.2 mg/ml was spun at 300 g for 30 s to remove large aggregates. 3 uL was added to the grids for 1 minute, before blotting with Whatman No1 filter paper and washing three times with 20 uL drops of ultrapure water. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Blotting time 3 s, waiting time 30 s Sample volume 3 uL, blot force 10. | ||||||||||||
| Details | 40 uL of a protein sample with an OD_280 of 0.88 were injected onto a Cytiva Superdex 200 Increase 3.2/300 size-exclusion chromatography column, equilibrated in 150 mM NaCl, 20 mM MES pH 7.0, 1 mM CaCl2, collecting 50 uL fractions. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)
































 Homo sapiens (human)
Homo sapiens (human)