+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rho-ATPgS-Psu complex I | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transcription termination / Phage inhibitor / GENE REGULATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated activation of host transcription / ATP-dependent activity, acting on RNA / helicase activity / DNA-templated transcription termination / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / ATP hydrolysis activity / RNA binding / ATP binding / cytosol Similarity search - Function | |||||||||
| Biological species |   Enterobacteria phage P4 (virus) Enterobacteria phage P4 (virus) | |||||||||
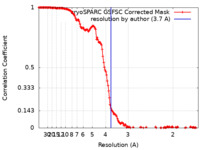
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Gjorgjevikj D / Wahl MC / Hilal T / Loll B | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Rho-ATPgS-Psu complex I Authors: Gjorgjevikj D / Wahl MC / Hilal T / Loll B | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17637.map.gz emd_17637.map.gz | 75.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17637-v30.xml emd-17637-v30.xml emd-17637.xml emd-17637.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17637_fsc.xml emd_17637_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_17637.png emd_17637.png | 72.8 KB | ||
| Filedesc metadata |  emd-17637.cif.gz emd-17637.cif.gz | 6.5 KB | ||
| Others |  emd_17637_half_map_1.map.gz emd_17637_half_map_1.map.gz emd_17637_half_map_2.map.gz emd_17637_half_map_2.map.gz | 318.7 MB 318.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17637 http://ftp.pdbj.org/pub/emdb/structures/EMD-17637 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17637 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17637 | HTTPS FTP |
-Validation report
| Summary document |  emd_17637_validation.pdf.gz emd_17637_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17637_full_validation.pdf.gz emd_17637_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_17637_validation.xml.gz emd_17637_validation.xml.gz | 24.1 KB | Display | |
| Data in CIF |  emd_17637_validation.cif.gz emd_17637_validation.cif.gz | 31.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17637 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17637 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17637 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17637 | HTTPS FTP |
-Related structure data
| Related structure data |  8peuMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17637.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17637.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_17637_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17637_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of E. coli transcription termination factor Rho with the ...
| Entire | Name: Complex of E. coli transcription termination factor Rho with the ATP analog, ATPgammaS and P4 polarity suppression protein Psu |
|---|---|
| Components |
|
-Supramolecule #1: Complex of E. coli transcription termination factor Rho with the ...
| Supramolecule | Name: Complex of E. coli transcription termination factor Rho with the ATP analog, ATPgammaS and P4 polarity suppression protein Psu type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: Transcription termination factor Rho
| Supramolecule | Name: Transcription termination factor Rho / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Polarity suppression protein
| Supramolecule | Name: Polarity suppression protein / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage P4 (virus) Enterobacteria phage P4 (virus) |
-Macromolecule #1: Transcription termination factor Rho
| Macromolecule | Name: Transcription termination factor Rho / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.070168 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNLTELKNTP VSELITLGEN MGLENLARMR KQDIIFAILK QHAKSGEDIF GDGVLEILQD GFGFLRSADS SYLAGPDDIY VSPSQIRRF NLRTGDTISG KIRPPKEGER YFALLKVNEV NFDKPENARN KILFENLTPL HANSRLRMER GNGSTEDLTA R VLDLASPI ...String: MNLTELKNTP VSELITLGEN MGLENLARMR KQDIIFAILK QHAKSGEDIF GDGVLEILQD GFGFLRSADS SYLAGPDDIY VSPSQIRRF NLRTGDTISG KIRPPKEGER YFALLKVNEV NFDKPENARN KILFENLTPL HANSRLRMER GNGSTEDLTA R VLDLASPI GRGQRGLIVA PPKAGKTMLL QNIAQSIAYN HPDCVLMVLL IDERPEEVTE MQRLVKGEVV ASTFDEPASR HV QVAEMVI EKAKRLVEHK KDVIILLDSI TRLARAYNTV VPASGKVLTG GVDANALHRP KRFFGAARNV EEGGSLTIIA TAL IDTGSK MDEVIYEEFK GTGNMELHLS RKIAEKRVFP AIDYNRSGTR KEELLTTQEE LQKMWILRKI IHPMGEIDAM EFLI NKLAM TKTNDDFFEM MKRS UniProtKB: Transcription termination factor Rho |
-Macromolecule #2: Polarity suppression protein
| Macromolecule | Name: Polarity suppression protein / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage P4 (virus) Enterobacteria phage P4 (virus) |
| Molecular weight | Theoretical: 21.393064 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MESTALQQAF DTCQNNKAAW LQRKNELAAA EQEYLRLLSG EGRNVSRLDE LRNIIEVRKW QVNQAAGRYI RSHEAVQHIS IRDRLNDFM QQHGTALAAA LAPELMGYSE LTAIARNCAI QRATDALREA LLSWLAKGEK INYSAQDSDI LTTIGFRPDV A SVDDSREK FTPAQNMIFS RKSAQLASRQ SV UniProtKB: Polarity suppression protein |
-Macromolecule #3: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 10 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 10 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: NITROGEN / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 6066 / Average exposure time: 40.57 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.9 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | Crystal structures of Rho (PDB ID: 1PV4) and Psu (PDB ID: 3RX6) were manually placed in the cryoEM reconstructions and each protomer was adjusted by rigid body fitting and segmental real-space refinement using Coot (version 0.9.6). The models were refined by iterative rounds of real space refinement in PHENIX (version 1.20_4459) and manual adjustment in Coot. | ||||||
| Refinement | Space: REAL / Target criteria: Cross-correlation coefficient | ||||||
| Output model |  PDB-8peu: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)